J Sci Discov (2017); 1(2):jsd17015; DOI:10.24262/jsd.1.2.17015; Received August 12nd, 2017, Revised September 24th, 2017, Accepted October 20th, 2017, Published October 29th,2017.
Microbial ecology of Scardovia wiggsiae-positive and negative samples
Amy Tam1, Karl Kingsley2, *
1Amy Tam, DDS is an Orthodontic Resident at the University of Nevada, Las Vegas – School of Dental Medicine, Department of Advanced Education Program in Orthodontics and Dentofacial Orthodpedics, 1700 W Charleston Avenue, Las Vegas, Nevada, 89106, USA, (702) 774-2690; Amy.Tam@sdm.unlv.edu
2Karl Kingsley, PhD, MPH is Director of Student Research and Professor of Biomedical Sciences at the University of Nevada, Las Vegas – School of Dental Medicine, Department of Biomedical Sciences, 1001 Shadow Lane, Las Vegas, Nevada, 89106, USA, (702) 774-2623; Karl.Kingsley@unlv.edu;
* Correspondence: Karl Kingsley, PhD, MPH, Department of Biomedical Sciences University of Nevada, Las Vegas – School of Dental Medicine, 1001 Shadow Lane,
Tel: (702) 774-2630, Fax: (702) 774-2721.E-mail: Karl.Kingsley@unlv.edu
Abstract
Recent studies have reported a novel cariogenic pathogen Scardovia wiggsiae among patients with poor oral health. The prevalence of this organism in the context of additional caries risk factors, such as Orthodontic treatment has yet to be fully explored. In addition, few studies have evaluated the presence of other pathogenic organisms with respect to Scardovia-positive patients. Recent studies at this institution have revealed S. wiggsiae among orthodontic and non-orthodontic patients have generated sufficient information to briefly summarize and characterize the microbial ecology of Scardovia wiggsiae-positive and negative saliva samples within this patient population. In brief, the cariogenic pathogens S. wiggsiae and S. mutans can be found separately on in combination in approximately half of all patient samples. However, the presence of other organisms, most notably Selenomonas noxia and Tannerella forsythia were only found in the Scardovia-negative samples – which may suggest these organisms or other factors that promote their growth or aggregation may selectively inhibit S. wiggsiae.
Keywords: Scardovia wiggsiae, Pediatric, Adult Saliva Screening, Microbial Ecology
Introduction
The studies from this institution were initiated to reveal whether samples from pediatric (and some adult) patients harbored DNA specific for Scardovia wiggsiae (S. wiggsiae) [1]. Although the first descriptions of this organism were from children with severe early childhood caries, the main finding from this initial pilot study was the discovery of S. wiggsiae in approximately one-quarter of both the pediatric and adult patient saliva samples [2,3]. A more recent study from this group confirmed these findings among a much larger sample of both pediatric and adult patients, further support for the growing evidence that Scardovia may be part of the oral microbial flora in patients with severe early childhood caries, as well as in pediatric and adult patients with other caries risk factors and profiles [4-6].
For example, some evidence has suggested S. wiggsiae may be a smaller part of the normal oral flora in patients without caries [7,8]. However, other studies have now demonstrated that orthodontic therapy may increase the risk of both caries and of high levels of Scardovia in some patients [9]. This observation has also been made in studies from this group, which has demonstrated the presence of this organism in nearly twice the percentage of pediatric orthodontic patients compared with either adult orthodontic patients or pediatric patients without orthodontic appliances [10]. In fact, two additional studies of orthodontic patients have recently been completed, which provide more support for these observations [11,12].
These studies provide the rationale for a more thorough investigation and screening of patient samples, which have been demonstrated to harbor S. wiggsiae [13,14]. Based upon these studies, sufficient data now exist to support the overall objective of this study, which was to provide a more detailed analysis and description of the microbial ecology found among Scardovia-positive and –negative patients samples.
Results
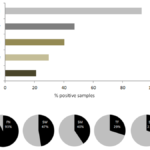
Using the previous microbial screening studies from this institution, results for the prevalence of several oral microbial species were compiled for analysis (Figure 1). These data clearly demonstrate that bridge species, such as Fusobacterium nucleatum (FN) are present in the overwhelming majority of samples – supporting other similar observations [15-17]. These data also demonstrate that both the cariogenic pathogens S. wiggsiae (SW) and Streptococcus mutans (SM) are found in nearly half of all patient samples [1,4,9,11-14]. However, other oral pathogens, such as T. forsythia (TF) and Selenemonas noxia (SN) were only found in approximately one-quarter of patient samples [13,18].
Figure 1. Combined analysis of institutional screening studies of oral microbial pathogens. An analysis of all patient saliva screening studies revealed most samples harbored DNA from the periodontal pathogen F. nucleatum (FN), while smaller subsets were found to contain the cariogenic pathogens S. wiggsiae (SW) and S. mutans (SM). Additional oral microbes T. forsythia (TF) and S. noxia (SN) were also present in approximately one-quarter of all samples analyzed.
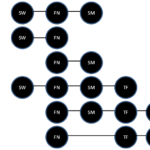
While these data provide some limited information regarding prevalence for several important oral species, more detailed analysis of data specific to each sample screened for multiple studies can provide a more comprehensive profile of the microbial flora (Figure 2). More specifically, a subset of samples that were screened in multiple studies provides a detailed analysis of the major pathogenic organisms, such as S. wiggsiae and S. mutans, which were present in nearly half of the same samples – either alone or in combination with F. nucleatum. These data clearly demonstrate that although S. wiggsiae and S. mutans are commonly found with F. nucleatum, they may also both be found concomitantly.
Figure 2. Comprehensive microbial oral patient sample profile. Most samples harbored the microbial bridge species F. nucleatum (FN), which was often found in combination with other oral pathogens. The cariogenic pathogens S. wiggsiae (SW) and S. mutans (SM) were found in combination with FN in approximately one-fourth of samples analyzed, and were also found to be present concomitantly in another subset (26.3%). Other organisms, such as T. forsythia (TF), were found in smaller subsets, while S. noxia (SN) was only found among the SW-negative samples.
Conclusions
These data may suggest that S. noxia and S. wiggsiae may occupy distinct, non-overlapping niches, which may differ significantly from the interactions observed with F. nucleatum. The limited numbers of studies available regarding S. wiggsiae prevalence have suggested that S. wiggsiae and S. mutans may inhabit similar and overlapping niches within the oral microbiome. In fact, studies now suggest the potential for both competition and interactive inhibition between these organisms within the oral cavity. The preliminary data from this pilot study suggest S. mutans and S. wiggsiae may, in fact, be present in some of the same patients and may not therefore be exclusively competitive – at least in this patient population. However, due to the large differences observed among these samples, further research will be needed to more fully elucidate these interactions and to explore the potential ramifications for oral microbial ecology and the implications for predictive saliva screening.
Conclusion
We conclude that Partial gastric outlet obstruction of unclear etiology might be attributed to duodenal lymphangectasia. So we recommend considering this pathology as one of the causes of partial gastric outlet obstruction. Moreover, we encourage reporting similar case scenarios so we will come to solid base to confirm our consideration.
Conflict of interest
None
Acknowledgments
The authors would like to thank the Department of Advanced Education Program in Orthodontics and Dentofacial Orthodpedics as well as Dr. Mobley and the Office of Research at the University of Nevada, Las Vegas – School of Dental Medicine for funding to support this project.
References
1. Catmull J, Row L, Repp MR, Heslington C, Miller T, Diamond J, Howard KM, Kingsley K. Newly identified cariogenic pathogen Scardovia wiggsiae detected by polymerase chain reaction in saliva of teenagers and adults in Southern Nevada. Forum for Dental Student Research and Innovation (FDSRI). 2014; (Spring); 22-29.
2. Tanner AC, Mathney JM, Kent RL, Chalmers NI, Hughes CV, Loo CY, Pradhan N, Kanasi E, Hwang J, Dahlan MA, Papadopolou E, Dewhirst FE. Cultivable anaerobic microbiota of severe early childhood caries. J Clin Microbiol. 2011; 49(4):1464-74.
3. Tanner AC, Kent RL Jr, Holgerson PL, Hughes CV, Loo CY, Kanasi E, Chalmers NI, Johansson I. Microbiota of severe early childhood caries before and after therapy. J Dent Res. 2011;90(11):1298-305.
4. Row L, Repp MR, Kingsley K. Screening of a pediatric and adult clinic population for the dental caries pathogen Scardovia wiggsiae using saliva. J Clin Pediatr Dent. 2016;40(6):438-444.
5. Richards VP, Alvarez AJ, Luce AR, Bedenbaugh M, Mitchell ML, Burne RA, Nascimento MM. Microbiomes of Site-Specific Dental Plaques from Children with Different Caries Status. Infect Immun. 2017;85(8): pii: e00106-17.
6. Eriksson L, Lif Holgerson P, Johansson I. Saliva and tooth biofilm bacterial microbiota in adolescents in a low caries community. Sci Rep. 2017;7(1):5861.
7. Tian L, Sato T, Niwa K, Kawase M, Mayanagi G, Washio J, Takahashi N. PCR-dipstick DNA chromatography for profiling of a subgroup of caries-associated bacterial species in plaque from healthy coronal surfaces and periodontal pockets. Biomed Res. 2016; 37(1):29-36.
8. Henne K, Rheinberg A, Melzer-Krick B, Conrads G. Aciduric microbial taxa including Scardovia wiggsiae and Bifidobacterium spp. in caries and caries free subjects. Anaerobe. 2015;35(Pt A):60-5.
9. BJ Streiff, M Seneviratne, K Kingsley. Screening and Prevalence of the Novel Cariogenic Pathogen Scardovia wiggsiae among Adult Orthodontic and Non-Orthodontic Patient Saliva Samples. Int J Dent Oral Health 2015; 1 (6). [Epub ahead of print]
10. Tanner AC, Sonis AL, Lif Holgerson P, Starr JR, Nunez Y, Kressirer CA, Paster BJ, Johansson I. White-spot lesions and gingivitis microbiotas in orthodontic patients. J Dent Res. 2012; 91(9):853-8.
11. Reyes N, Pollock A, Whiteley A, Kingsley K, Howard KM. Prevalence of Scardovia wiggsiae among a pediatric Orthodontic patient population. EC Dental Science. 2017; 13(5): 203-210.
12. Milne W, Rezeai G, Whiteley A, Kingsley K. Cariogenic pathogen Scardovia wiggsiae screening among pediatric orthodontic patients: A pilot study. Current Research in Dentistry. 2017. In Press.
13. McDaniel S, McDaniel J, Tam A, Kingsley K, Howard KM. Oral Microbial Ecology Of Selenomonas Noxia And Scardovia Wiggsiae. Microbiology Research Journal International. 2017; 21(3): 1-8.
14. McDaniel J, McDaniel S, Tam A, Kingsley K, Howard KM. Scardovia wiggsiae and Streptococcus mutans Prevalence Among Dental School Patients: A pilot study. Journal of Advances in Microbiology. 2017; 5(1): 1-8.
15. Boutin S, Hagenfeld D, Zimmermann H, El Sayed N, Höpker T, Greiser HK, Becher H, Kim TS, Dalpke AH. Clustering of Subgingival Microbiota Reveals Microbial Disease Ecotypes Associated with Clinical Stages of Periodontitis in a Cross-Sectional Study. Front Microbiol. 2017; 8:340.
16. Lima BP, Shi W, Lux R. Identification and characterization of a novel Fusobacterium nucleatum adhesin involved in physical interaction and biofilm formation with Streptococcus gordonii. Microbiologyopen. 2017; 6(3).
17. Jolley D, Wonder K, Chang E, Kingsley K. Oral microbial prevalence of periodontal pathogens among orthodontic patients. International Journal of Dentistry and Oral Health (IJDOH) 2016; 1(6).
18. Bui Q, Nguyen C, McDaniel J, McDaniel S, Kingsley K, Howard KM. Selenomonas noxia screening among pediatric patient samples: a pilot study. J Oral Heal Dent Car. 2017; 1:1009.
Copyright
© This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/