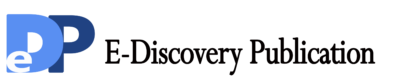J Med Discov (2020); 5(3):jmd20035; DOI:10.24262/jmd.5.3.20035; Received April 28th, 2020, Revised July 12nd, 2020, Accepted August 03rd, 2020, Published August 22nd, 2020.
Increased expression of TBL1XR1 is associated with poor prognosis in patients with clinical N0 tongue squamous cell cancer
Zhen Liao1,2,Yan-Ling Liu1,2,Shi-Min Zhuang3
1Dept of Operation Theater Services, Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center of Cancer Medicine, 651 Dongfeng Dong Road, Guangzhou 510060, People’s Republic of China
2Dept of Intensive Care Unit, Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center of Cancer Medicine, 651 Dongfeng Dong Road, Guangzhou 510060, People’s Republic of China.
3Dept of Otolaryngology-Head & Neck Surgery, The Sixth Affiliated Hospital of Sun Yat-sen University,26 Yuanchun Two Road, Guangzhou, Guangdong, 510655,People’s Republic of China.
* Correspondence:Shi-Min Zhuang, Dept of Otolaryngology-Head & Neck Surgery, The Sixth Affiliated Hospital of Sun Yat-sen University,26 Yuanchun Two Road, Guangzhou, Guangdong, 510655,People’s Republic of China.
Abstract
Transducin β-like 1 X-linked receptor 1 (TBL1XR1) overexpression was associated with the progression of several solid tumors, with resulted in adverse clinical outcomes. The study aimed to determine the expression and prognostic of TBL1XR1 in clinical N0 tongue squamous cell cancer (cN0 TSCC). TBL1XR1 expression was examined in 8 paired samples of TSCC and adjacent non-cancerous tongue tissues, and 4 TSCC lines and 1 normal oral keratinocytes (NOK). TBL1XR1 expression levels were also assessed in 174 TSCC samples. The association of TBL1XR1 expression with clinicopathological features and clinical outcome was analyzed. Analysis revealed TBL1XR1 expression was significantly elevated in cN0 TSCC tissue compared with the matched adjacent non‑cancerous tongue tissue,and elevated in TSCC lines compared with that of NOK. In addition, the results demonstrated TBL1XR1 expression was positively correlated with the pathological stage (P=0.001), T classification (P=0.004) and N classification (P=0.018). Kaplan-Meier analysis found cN0 TSCC patients with increased TBL1XR1 expression have shorter overall survival time and higher recurrence rates. TBL1XR1 expression was an independent factor for predicting the poor overall survival of TSCC patients. In conclusion, our study demonstrated high TBL1XR1 levels is associated with poor prognosis in patients with TSCC, may be valuable for the clinical evaluation of high-risk patients.
Keywords: TBL1XR1, tongue squamous cell cancer, prognosis
Introduction
Tongue cancer is one of the most common and lethal oral cancer, the most common pathological type of oral tongue cancer is squamous cell carcinoma[1, 2]. Despite advances in standard treatment strategies (e.g.surgery, chemotherapy, and radiotherapy), the 5-year survival rate in patients with tongue squamous cell cancer(TSCC) has remained high [3], 78 % for cancer exhibiting local spread, 63 % for cancer exhibiting regional spread, and 36 % for cancer exhibiting distant spread [4, 5].
Though aggressive management may reduce the recurrence rate, result in potentially devastating consequences on quality of life[6,7]. Clinical evidences indicate that metastasis is the most important poor prognostic factors for patient diagnosis with TSCC [8]. Tumorigenesis is a complex multi-step process characterized by uncontrolled cell growth and tumor formation. Therefore, it is of great value to identify the high-risk patients who may benefit from more aggressive primary surgery or adjuvant treatment following surgery.
Reports have shown that tumorigenesis is associated with the progressive accumulation of genetic and epigenetic alterations in genes and proteins that regulate cell proliferation, cell death and genomic instability[9,10]. Increasing evidences have recognized that the epithelial to mesenchymal transition (EMT), a driver ofinvasion and metastasis of cancer, may play a pivotal role in multiple types of tumor cell metastatic dissemination by endowing cells with a more motile, invasive potential[11, 12, 13, 14].
Previous studies have established TBL1XR1 as a key player in the regulation of multiple signaling pathways (Wnt/β-catenin, Notch, NF-κB, and nuclear receptor) and gene transcription, which all involved in EMT [15, 16, 17, 18].
TBL1XR1 was originally identified as a component of the nuclear receptor corepressor complex [19], In addition, TBL1XR1 has been found to affect carcinogenesis and tumor progression in several solid tumors, including lung cancer [20], breast cancer [22, 22], live cancer [23,24], prostate cancer[25], colorectal cancer [18] and nasopharyngeal carcinoma [26].
Based on above materials, we have evidences to assume TBL1XR1 promoted the progression and invasiveness of cancer. But the clinical significance and biological function of TBL1XR1 in the progression of tongue squamous cancer remain poorly understood. In this study, we found that TBL1XR1 was significantly upregulated in tongue squamous cells and tissues, and the expression was closely correlated with clinicopathologic features in patients with tongue squamous cancer. Furthermore, we have demonstrated that TBL1XR1 plays an important role in occurrence and progress of tongue squamous cancer and require next step thorough research.
Materials and Methods
Patients and tissue specimens
For the use of these clinical materials for research purposes, prior patient consent and approval from the Ethics Committees of the Cancer Center, Sun Yat-sen University in advance of the study. For RT-PCR and Western blotting analysis, eight matched pairs of tumors tissue and adjacent noncancerous tissue samples were obtained from glossectomy specimens of patients diagnosed with cN0 tongue squamous cancer immediately after surgery and stored at −80°C. A total of 179 individual paraffin-embedded cN0 tongue cancer samples, which were histopathologically and clinically diagnosed tongue cancer patients treated at the Cancer Center, Sun Yat-sen University between 2000 and 2005. Clinical follow-up data was available for a minimum of 5 years or until death. The clinical information this patient cohort is summarized in Table 1.
Cell lines
The TSCC cell lines, CAL27, SCC-25 and SCC-9, were purchased from American Type Culture Collection (Manassas, VA, USA). The TSCC cell lines, CAL33, and normal oral keratinocytes (NOK) were kindly provided by J. Silvio Gutkind (NIH, Besthesda, MD). CAL27, CAL33, SCC-9 and SCC-25 were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Inc., Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS). NOK cells were maintained in KSFM (Gibco, USA). Cells were incubated in a 5% CO2 atmosphere at 37°C.
Table 1. Clinicopathologic characteristics and TBL1XR1 expression of patients with clinically N0 oral tongue cancer of the study cohort (n=174)
| Characteristics | Number of cases (%) |
| Gender | |
| Male | 105 (60.3) |
| Female | 69 (39.7) |
| Age (years) | |
| < 53 | 88 (50.6) |
| ≥ 53 | 86 (49.4) |
| Pathologic stage | |
| Ⅰ | 78 (44.8) |
| Ⅱ | 64 (36.8) |
| Ⅲ | 17 (9.8) |
| Ⅳ | 15 (8.6) |
| T classification | |
| T1 | 85 (48.9) |
| T2 | 84 (48.3) |
| T3 | 5 (2.8) |
| N classification | |
| N0 | 144 (82.8) |
| N1 | 15 (8.6) |
| N2 | 15 (8.6) |
| Pathologic differentiation | |
| Well | 136 (76.0) |
| Moderately | 36 (20.1) |
| Poorly | 7 (3.9) |
| Recurrence | |
| No | 93 (53.4) |
| Yes | 81 (44.6) |
| Vital status (at follow-up) | |
| Alive | 60 (34.5) |
| Dead | 114 (65.5) |
| Expression of GOLPH3 | |
| Low or none expression | 55(31.6) |
| High expression | 119(68.4) |
Real-time PCR (RT-PCR)
Total RNA from cultured cells and fresh tissues were extracted using the Trizol reagent (Invitrogen) according to the manufacturer’s instruction. The extracted RNA was pretreated with RNase-free DNase, and about 2ug RNA from each sample was used for cDNA synthesis with iScriptcDNA Synthesis Kit (Bio-Rad Laboratories,Hercules, CA). Real-time PCR primes were designed using the Primer Express Software Version 2.0 and the primer sequences are: TBL1XR1 forward primer: GAATTTCCTTGTGCCTCCAT; TBL1XR1 reverse primer: TGCAACTGAATATCCGGTCA;Glyceraldehyde-3-phos hate dehydrogenase (GAPDH) forward prime: 5′-GACTCATGACCACAGTCC ATGC-3′; GAPDH reverse primer: 5′-AGAGGCA GGGATGATGTTCTG-3′. Expression data were normalized to the geometric mean of housekeeping gene GAPDH to control the variability in expression levels.
Western blotting
Western blot analysis was performed according to standard methods as described previously [27], using anti-TBL1XR1rabbit polyclonal antibody (1:3,000; Sigma). Blot membranes were stripped and reprobed with anti-GAPDH antibody (1:1,000; Sigma) as a loading control.
Immunohistochemistry
Immunohistochemistry methods and scoring for GOLPH3 expression were done as previously described[27]. Two independent pathologists blinded to the clinical parameters conducted the staining index (SI) for TBL1XR1 expression. The staining results were scored based on the following criteria: (i) percentage of positive tumor cells in the tumor tissue: 0 (0%), 1 (1%–25%), 2 (26%–50%), 3 (51%–75%) and 4 (76%–100%); (ii) staining intensity: 0 (no staining), 1 (weak staining = light yellow), 2 (moderate staining = yellow brown), and 3 (strong staining = brown). The SI was calculated as staining intensity score × proportion of positive tumor cells (range from 0 to 12) [34]. An optimal cutoff value was identified: the SI > 6 was used to define as TBL1XR1 high expression while SI ≤ 6 as TBL1XR1 low expression.
Statistical analyses
All statistical analyses were carried out using the SPSS version 17.0 statistical software packages. The correlation between TBL1XR1 expression and the clinicopathological characteristics was analyzed by the chi-square test. Survival curves were plotted by the Kaplan-Meier method and compared with the log-rank test. Multivariate survival analysis was performed for all parameters found to be significant in the univariate analysis using the Cox regression model. A two-sided probability value of less than 0.05 was considered to be statistically significant.
Results
TBL1XR1 is overexpressed in oral tongue cancer cell lines
To investigate the potential role of TBL1XR1 in the tumorigenesis of oral tongue cancer, the expression of TBL1XR1 mRNA and protein were determined for four oral tongue cancer cell lines (SCC-25, SCC-9, CAL27 and CAL33) and compared with TBL1XR1 expression in normal oral keratinocytes (NOK). TBL1XR1 mRNA expression was higher in oral tongue cancer cell lines than that in NOK (Figure 1A), and GOLPH3 protein was highly expressed in oral tongue cancer cell lines and only weakly expressed in TEC (Figure 1B).
Figure 1. Increased expression of TBL1XR1 protein in tongue squamous cancer cell lines (A and B) Expression of TBL1XR1 mRNA and protein in tongue cancer cell lines (SCC-9, SCC-25, CAL27, CAL33, Tca-8113) and normal oral keratinocytes (NOK) were examined by RTPCR (A) and Western blotting (B). Expression levels were normalized to GAPDH. Error bars represent standard deviation of the mean (SD) calculated from three parallel experiments.
TBL1XR1 is overexpressed in oral tongue cancer tissues
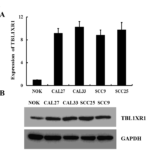
To investigate TBL1XR1 mRNA and protein expression in cN0 oral tongue cancer, RT-PCR and Western blotting analyses were done on eight matched pairs of oral tongue cancer samples (T) and adjacent noncancerous tissue samples (ANT). TBL1XR1 mRNA was expressed at higher levels in all oral tongue cancer tissue samples than that in adjacent noncancerous tissues (Figure 2A). Consistent with this data, TBL1XR1 protein was also up-regulated in cN0 oral tongue cancers compared with the matched controls (Figure 2B).
Figure 2. Increased expression of TBL1XR1 protein in tongue squamous cell cancer tissue (A) Average T/N ratios of TBL1XR1 mRNA expression in paired tongue cancer (T) and adjacent noncancerous tissues (N) was quantified by RTPCR and normalized to GAPDH. Error bars represent standard deviation of the mean (SD) calculated from three parallel experiments. (B) Western blot analysis of TBL1XR1 protein expression in four matched pairs of oral tongue cancer (T) and adjacent noncancerous tissues (N). GAPDH was the loading control.
TBL1XR1 overexpression is associated with clinical features of cN0 oral tongue cancer
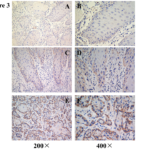
We further investigated the link between TBL1XR1 protein expression and the clinicopathological characteristics of oral tongue cancer using a panel of 174 paraffin-embedded, archived oral tongue cancer specimens. The clinicopathological characteristics were shown in Table 1. 167 of the total 174 oral tongue cancers (95.9%) were positive for TBL1XR1 based on immunohistochemical staining. High TBL1XR1 protein expression was detected in 119 samples (68.4%) and weak or negative staining was observed in 55 tumor samples (31.6%, Figure 3). Statistical analysis showed a strong correlation between TBL1XR1 expression and clinicopathological characteristics of cN0 oral tongue cancer, including pathologic stage (P =0.001), T classification (P = 0.004), N classification (P =0.018), and recurrence (P =0.001). In contrast, TBL1XR1 expression had no correlation with age, gender and tumor differentiation (Table 2).
Figure 3. Immunohistochemical of TBL1XR1 in tongue squamous cell cancer sections. Representative immunohistochemical images of cN0 oral tongue cancer tissue specimens indicating strong TBL1XR1 staining (A and B); moderate TBL1XR1 staining (C and D); and weak TBL1XR1 staining (E and F). Magnification is ×200 (A, C and E) or ×400 (B, D and F).
Table 2. Correlation between TBL1XR1 expression and clinicopathologic characteristics of patients with clinically N0 oral tongue cancer
| Characteristics | TBL1XR1 expression | |||
| n |
Low or none, n. (%) |
High, n. (%) |
χ2 test P | |
| Gender | 0.097 | |||
| Male | 105 | 28 (26.7) | 77 (73.3) | |
| Female | 69 | 27 (39.1) | 42 (60.9) | |
| Age (years) | 0.331 | |||
| < 53 | 88 | 31 (35.2) | 57 (64.8) | |
| ≥ 53 | 86 | 24 (27.9) | 62 (72.1) | |
| Pathological stage | 0.001 | |||
| Ⅰ | 78 | 37 (47.4) | 41 (52.6) | |
| Ⅱ | 64 | 15 (23.4) | 49 (76.6) | |
| Ⅲ | 17 | 1 (5.9) | 16 (94.1) | |
| Ⅳ | 15 | 2 (13.3) | 13 (86.7) | |
| T classification | 0.004 | |||
| T1 | 85 | 37 (43.5) | 48 (56.5) | |
| T2 | 84 | 17 (20.2) | 67 (79.8) | |
| T3 | 5 | 1 (20) | 4 (80) | |
| N classification | 0.018 | |||
| N0 | 144 | 52 (36.1) | 92 (63.9) | |
| N1 | 15 | 1 (6.7) | 14 (93.3) | |
| N2 | 15 | 2 (13.3) | 13 (86.7) | |
| Pathologic differentiation | 0.189 | |||
| Well | 131 | 45 (34.4) | 86 (65.6) | |
| Moderately | 36 | 7 (19.9) | 29 (80.1) | |
| Poorly | 7 | 3 (42.9) | 4 (57.1) | |
| Recurrence | 0.001 | |||
| No | 93 | 42 (45.2) | 51 (54.8) | |
| Yes | 81 | 13 (26.0) | 68 (84.0) | |
| Vital status (at follow-up) | 0.001 | |||
| Alive | 114 | 48 (42.1) | 66 (57.9) | |
| Dead | 60 | 7 (11.7) | 53 (88.3) | |
Association between TBL1XR1 expression and patient survival
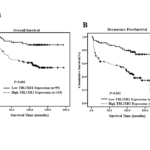
Patient survival analysis showed a clear negative correlation between the level of TBL1XR1 protein expression and both the overall survival and recurrence -free survival of patients with cN0 oral tongue cancer (P = 0.001 and 0.001, respectively; Figure 4A, B). The cumulative 5-year overall and recurrence -free survival rates for patients with high levels of TBL1XR1 expression were 59% and 41%, whereas for patients with low TBL1XR1 expression the rates were 84% and 67%, respectively. Cox regression revealed that N classification (relative risk, 1.659, CI: 1.201–2.293, P = 0.002), pathologic differentiation degree (relative risk, 2.040, CI: 1.379–3,020, P = 0.001) and TBL1XR1 overexpression (relative risk, 3.804, CI: 1.692–8.552, P = 0.001) were independent prognostic factors for poor overall survival.
Figure 4. TBL1XR1 affects overall survival and disease-free survival. Kaplan–Meier curves with univariate analysis (log-rank) for cN0 oral tongue cancer patients with high TBL1XR1 expression (n=119) versus low or no TBL1XR1 expression (n=55) for overall survival (A) and disease-free survival (B).
Discussion
The pivotal finding of this study is that the elevated expression of TBL1XR1 correlated with a poor prognosis and reduced survival of TSCC patients. Furthermore, Cox regression model indicated that TBL1XR1was an independent prognostic factor. These findings provide evidence that upregulation of TBL1XR1 plays an important role in promoting progression of tongue squamous cancer.
As the most common cancer diagnosed in the oral cavity, the poor prognosis of oral tongue cancer is mainly a consequence of its unusual histological makeup, which makes it poorly equipped to resist invasion and metastasis [28, 29].
In clinical practice, TNM classification system, the most pervasive index for developing treatment protocols and predicting clinical outcome, is not sufficiently reliable for predicting clinical outcome or for providing detailed information on the biological characteristics of a malignancy [30, 31].
TBL1XR1 is a component of SMRT/NCoR corepressor complex and involves in the exchange between corepressor and coactivators complexes by unliganded nuclear receptors [32, 33].
TBL1XR1 has been confirm to affect carcinogenesis and tumor progression in several solid tumors, including lung cancer [20], breast cancer [21, 22], live cancer [23, 24], prostate cancer [25], colorectal cancer [26] and nasopharyngeal carcinoma [27]. TBL1XR1 participate in several signal paths to mediate EMT, which is the main mechanism for invasion and metastasis [18, 22, 26]. Consistent with these results, our data showed that expression of TBL1XR1 was elevated in tongue squamous cell lines and tissue. We have evidences to assume TBL1XR1 promoted the progression and invasiveness of tongue squamous cancer. According to our clinical experience, the patients with poor pathologic differentiation or N+ stage should receive adjuvant treatment following surgery. Our results reconfirm the role of pathologic differentiation or N stage for developing treatment protocols and predicting clinical outcome. Elevated levels of TBL1XR1 protein positively correlated with pathological stage, T classification, N classification. Moreover, cN0 oral tongue cancer patients with elevated TBL1XR1 expression had significantly shorter overall and recurrence-free survival time than patients with lower TBL1XR1 expression. We therefore report that TBL1XR1 is a risk factor for cN0 oral tongue cancer. Thus, the detection of overexpressed TBL1XR1 in cN0 oral tongue cancer should identify high-risk tumor phenotypes that require more aggressive primary surgery or adjuvant treatment following surgery.
Although several new understandings of TBL1XR1in tongue squamous cancer are provided in this study. The underlying mechanism of TBL1XR1-mediated tongue cancer progression, the role of TBL1XR1 in malignant transformation and cell growth and its effects on clinical outcome remain to be defined. The predictive value of TBL1XR1 was found by a retrospective study, which is less convincing. But the mechanism and the prospective study are required to stepwise research.
Conclusion
In conclusion, our studies offer some insight into the function of TBL1XR1 in tongue squamous cell carcinoma. We suggest that determining TBL1XR1 expression levels in cN0 oral tongue cancers may help to identify patients harboring occult micrometastases that require more aggressive treatment and may therefore complement the current TNM classification to enable better risk stratification and election for adjuvant therapy. We further propose that targeting TBL1XR1 may be a useful strategy for developing novel therapeutic modalities.
Conflict of interest
None
Acknowledgments
None
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29.
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011;61:69-90.
- Marsh D, Suchak K, Moutasim KA, Vallath S, Hopper C, Jerjes W, Upile T, Kalavrezos N, Violette SM, Weinreb PH, et al. Stromal features are predictive of disease mortality in oral cancer patients. J Pathol. 2011;223(4):470-81.
- Liao CT, Kang CJ, Lee LY, Hsueh C, Lin CY, Fan KH, Wang HM, Ng SH, Lin CH, Tsao CK, et al. Association between multidisciplinary team care approach and survival rates in patients with oral cavity squamous cell carcinoma. Head Neck 2016;38 Suppl 1:E1544-E1553.
- Choi SW, Moon EK, Park JY, Jung KW, Oh CM, Kong HJ, Won YJ. Trends in the incidence of and survival rates for oral cavity cancer in the Korean population. ORAL DIS 2014;20:773-779.
- van Nieuwenhuizen AJ, Buffart LM, Brug J, Leemans CR, Verdonck-de LI. The association between health related quality of life and survival in patients with head and neck cancer: A systematic review. ORAL ONCOL 2015;51:1-11.
- Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. LANCET 2008;371:1695-1709.
- Fusco A, Fedele M. Roles of HMGA proteins in cancer. NAT REV CANCER 2007;7:899-910.
- Ponder BA. Cancer genetics. NATURE 2001;411:336-341.
- Bartek J. DNA damage response, genetic instability and cancer: From mechanistic insights to personalized treatment. MOL ONCOL 2011;5:303-307.
- Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH, et al. EMT and dissemination precede pancreatic tumor formation. CELL 2012;148:349-361.
- Kalluri R. EMT: When epithelial cells decide to become mesenchymal-like cells. J CLIN INVEST 2009;119:1417-1419.
- Morishita A, Zaidi MR, Mitoro A, Sankarasharma D, Szabolcs M, Okada Y, D’Armiento J, Chada K. HMGA2 is a driver of tumor metastasis. CANCER RES 2013;73:4289-4299.
- Waerner T, Alacakaptan M, Tamir I, Oberauer R, Gal A, Brabletz T, Schreiber M, Jechlinger M, Beug H. ILEI: A cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. CANCER CELL 2006;10:227-239.
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. CELL 2004;116:511-526.
- Choi HK, Choi KC, Yoo JY, Song M, Ko SJ, Kim CH, Ahn JH, Chun KH, Yook JI, Yoon HG. Reversible SUMOylation of TBL1-TBLR1 regulates beta-catenin-mediated Wnt signaling. MOL CELL 2011;43:203-216.
- Hoberg JE, Yeung F, Mayo MW. SMRT derepression by the IkappaB kinase alpha: A prerequisite to NF-kappaB transcription and survival. MOL CELL 2004;16:245-255.
- Li J, Wang CY. TBL1-TBLR1 and beta-catenin recruit each other to Wnt target-gene promoter for transcription activation and oncogenesis. NAT CELL BIOL 2008;10:160-169.
- Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. MOL CELL 2002;9:611-623.
- Liu Y, Sun W, Zhang K, Zheng H, Ma Y, Lin D, Zhang X, Feng L, Lei W, Zhang Z, et al. Identification of genes differentially expressed in human primary lung squamous cell carcinoma. LUNG CANCER 2007;56:307-317.
- Kadota M, Sato M, Duncan B, Ooshima A, Yang HH, Diaz-Meyer N, Gere S, Kageyama S, Fukuoka J, Nagata T, et al. Identification of novel gene amplifications in breast cancer and coexistence of gene amplification with an activating mutation of PIK3CA. CANCER RES 2009;69:7357-7365.
- Li X, Liang W, Liu J, Lin C, Wu S, Song L, Yuan Z. Transducin (beta)-like 1 X-linked receptor 1 promotes proliferation and tumorigenicity in human breast cancer via activation of beta-catenin signaling. BREAST CANCER RES 2014;16:465.
- Kulozik P, Jones A, Mattijssen F, Rose AJ, Reimann A, Strzoda D, Kleinsorg S, Raupp C, Kleinschmidt J, Muller-Decker K, et al. Hepatic deficiency in transcriptional cofactor TBL1 promotes liver steatosis and CELL METAB 2011;13:389-400.
- Kuang X, Zhu J, Peng Z, Wang J, Chen Z. Transducin (Beta)-Like 1 X-Linked Receptor 1 Correlates with Clinical Prognosis and Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma. Dig Dis Sci 2016;61:489-500.
- Daniels G, Li Y, Gellert LL, Zhou A, Melamed J, Wu X, Zhang X, Zhang D, Meruelo D, Logan SK, et al. TBLR1 as an androgen receptor (AR) coactivator selectively activates AR target genes to inhibit prostate cancer growth. Endocr Relat Cancer 2014;21:127-142.
- Chen SP, Yang Q, Wang CJ, Zhang LJ, Fang Y, Lei FY, Wu S, Song LB, Guo X, Guo L. Transducin beta-like 1 X-linked receptor 1 suppresses cisplatin sensitivity in nasopharyngeal carcinoma via activation of NF-kappaB pathway. MOL CANCER 2014;13:195.
- Li H, Guo L, Chen SW, Zhao XH, Zhuang SM, Wang LP, Song LB, Song M. GOLPH3 overexpression correlates with tumor progression and poor prognosis in patients with clinically N0 oral tongue cancer. J TRANSL MED 2012;10:168.
- Lim MS. Re: Correlational of oral tongue cancer inversion with matrix metalloproteinases (MMPs) and vascular endothelial growth factor (VEGF) expression, by Kim S-H, Cho NH, Kim K, et al. J SURG ONCOL 2006;93:253-254.
- Manning BD, Cantley LC. AKT/PKB signaling: Navigating downstream. CELL 2007;129:1261-1274.
- D’Cruz AK, Siddachari RC, Walvekar RR, Pantvaidya GH, Chaukar DA, Deshpande MS, Pai PS, Chaturvedi P. Elective neck dissection for the management of the N0 neck in early cancer of the oral tongue: Need for a randomized controlled trial. Head Neck 2009;31:618-624.
- Hayry V, Makinen LK, Atula T, Sariola H, Makitie A, Leivo I, Keski-Santti H, Lundin J, Haglund C, Hagstrom J. Bmi-1 expression predicts prognosis in squamous cell carcinoma of the tongue. Br J Cancer 2010;102:892-897.
- Tomita A, Buchholz DR, Obata K, Shi YB. Fusion protein of retinoic acid receptor alpha with promyelocytic leukemia protein or promyelocytic leukemia zinc finger protein recruits N-CoR-TBLR1 corepressor complex to repress transcription in vivo. J BIOL CHEM 2003;278:30788-30795.
- Y Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, Qin J, Wong J. Purification and functional characterization of the human N-CoR complex: The roles of HDAC3, TBL1 and TBLR1. EMBO J 2003;22:1336-1346.
Copyright
© This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/