J Med Discov (2018);3(3):jmd18036; DOI:10.24262/jmd.3.3.18036; Received August 9th, Revised August 23rd, Accepted August 25th, Published August 30th
Structure and Mechanogating Mechanism of Channel Piezo1 with Agonist
Shaopeng Chi1,*
1School of Pharmaceutical Sciences, Tsinghua-Peking Joint Center for Life Sciences, IDG/McGovern Brain Research Institute, Tsinghua University, Haidian District, Beijing, China.
* Correspondence: Shaopeng Chi, School of Pharmaceutical Sciences, Tsinghua-Peking Joint Center for Life Sciences, IDG/McGovern Brain Research Institute, Tsinghua University, Haidian District, Beijing, China. E-mail: csp16@mails.tsinghua.edu.cn.
Abstract
The mechanosensitive mouse Piezo1 channel function as key eukaryotic mechanotransducers. However, the structure and mechanogating mechanism of the mechanosensitive mouse Piezo1 channel with small molecule agonist remain unknown. New studies have now provided critical insights into the high-resolution cryo-electron microscopy structure and mechano-gating of the mechanosensitive mPiezo1 channel with agonist.
Keywords: Piezo1 channel, protein structure, mechanogating.
Introduction
Piezo family of genes are mechanically activated Ca2+-permeable non-selective cationic channels that mediate touch perception, proprioception, vascular development and brain development. They have been established as the long-sought-after mechanosensitive cation channels in mammals (1). In light of their physiological importance, both loss-of-function and gain-of-function mutations of Piezo family of genes have been associated with disease such as xerocytosis and lymphatic dysplasia (2, 3). Mammalian Piezos are large transmembrane proteins that are composed of about 2547 amino acids with 38 transmembrane-helix topology. Mouse Piezo1 has been previously determined, revealing its three-bladed, propeller-like trimeric architecture comprising a central pore module and three extended peripheral wings (4). The complex mPiezo1 protein might be divided into the peripheral blades and a central cation selective pore module (7). These features make structure and function studies of Piezo proteins highly challenging, partly explaining the slow progress in our understanding of themolecular basis of mechannogating properties of this novel of ion channels. In the latest researchs have now made major breakthroughs in demystifying Piezo channels.
In the first latest research, the present study show that the three-bladed, propeller-like cryo-EM structures of mPiezo1 and functionally reveal its mechanotransduction components. The mPiezo1 protein comprises 9 repetitive units of 4 transmembrane helices segments, assembling into the highly curved peripheral blade-like structure, which is critical for mechano-sensing and transduction (8). The last transmembrane encloses a hydrophobic pore, followed by three intracellular fenestration sites and side portals comprising pore-property-determining residues. The central region that forms a ~90 Å-long intracellular beam-structure, which bridges the blade to the pore. These results indicate that Piezo1 possesses an unprecedented 38-transmembrane topology and designated mechanotransduction components, enabling a lever-like apparatus for effectively transducing force from the mechanosensing blade to the ion-conducting pore, enabling long-distance mechanogating mechanism.
In the second latest research, the present study show that a novel Piezo1 chemical activators(Jedi) that directly bind to Piezo1 and modulate its mechanosensitivity and ion selectivity. Jedi act through the N-terminal extracellular loop regions that might reside in the distal blade-structure, activates Piezo1 through the extracellular side of the blade instead of the C-terminal ion-conducting pore, suggesting a long-range allosteric gating. Remarkably, Jedi-induced activation of Piezo1 requires the key mechanotransduction components, including the two extracellular loops( L15-16 and L19-20 ) in the distal transmembrane helical units or the mutating single(L1342/L1345) residues in the proximal end of the beam(Wang et al., 2018). The research further demonstrate that Jedi1 and Yoda1 (Yoda1 is a useful pharmacological tool(5) act through distinct mechanisms to modulate Piezo1. Piezo1 chemical activators (Jedi and Yoda1) appear to activate and modulate Piezo1 via acting on different loci along the blade-beam gating pathway. The the extracellular loops ( L15-16 and L19-20 ) mutants completely lose their ability to Jedi1/2, but remained Yoda1-induced a preferentially potentiated their poking currents , but not the stretching currents. In contrast, the L1342/L1345 mutants are insensitive to both Jedi and Yoda1. The data suggest that Jedi1/2 act on the upstream blade, while Yoda1 acts at the downstream beam. These results indicate that Piezo1 utilizes the beam as a lever-like apparatus for effectively transducing force from the mechanosensing blade to the ion-conducting pore, enabling long-distance mechanogating.
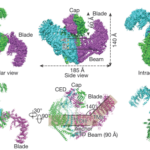
Fig 1. cryo-electron microscopy reconstruction of mouse Piezo1. a. Atomic model of the mPiezo1 ion channel, viewed from the top and the side. b. Cartoon models with each subunit colour-coded. Illustration adopted from
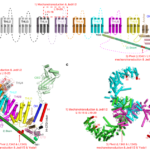
Fig.2. Topological and structural illustration of the designated mechanical and chemical activation of mouse Piezo1. a. topological model. b. structural model (from the reported cryo-EM structure of Piezo1) c. Top view of the trimeric mouse Piezo1 channel. Illustration adopted from
Conclusion and Perspective
Piezo proteins serve as a principal type of mechanotransduction channels. The cryo-electron microscopy structure of mouse Piezo1 and functionally shows that it forms a three-bladed, propeller shaped trimeric complex. The research identify 9 repetitive units consisting of 4 transmembrane helices each, which assemble into a highly curved blade-like structure. The mPiezo1 structure destribed represents the first high-resolution of this structurally and functionally novel of channel. Toward exactly understanding the structure-function relationship of this sophisticated type of mechanosensitive channels, future efforts should also be oriented toward obtaining Piezo structures in high-resolution, and making depth structure functional characterizations. The present disclosure relates to a bio-medicine, and more particularly to use of regulator used for activating Piezo in preparation of a medicament.
Conflict of interest
None
Acknowledgments
S. C. wrote the manuscript.
References
1. Coste B, Mathur J, Schmidt M, Earley T, Ranade S, Petrus M et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010; 330: 55-60.
2. Demolombe S, Duprat F, Honore E and Patel A. Slower Piezo1 inactivation in dehydrated hereditary stomatocytosis (xerocytosis). Biophys J. 2013; 105: 833-834.
3. Fotiou E, Martin-Almedina S, Simpson M, Lin S, Gordon K, Brice G et al. Novel mutations in PIEZO1 cause an autosomal recessive generalized lymphatic dysplasia with non-immune hydrops fetalis. Nat Commun. 2015; 6: 8085.
4. Ge J, Li W, Zhao Q, Li N, Chen M, Zhi P et al. Architecture of the mammalian mechanosensitive Piezo1 channel. Nature. 2015; 527: 64-69.
5. Syeda R, Xu J, Dubin A, Coste B, Mathur J, Huynh T et al. Chemical activation of the mechanotransduction channel Piezo1. 2015. Elife. 4.
6. Wang Y, Chi S, Guo H, Li G, Wang L, Zhao Q et al. A lever-like transduction pathway for long-distance chemical- and mechano-gating of the mechanosensitive Piezo1 channel. Nat Commun. 2018; 9: 1300.
7. Zhao Q, Wu K, Geng J, Chi S, Wang Y, Zhi P et al. Ion Permeation and Mechanotransduction Mechanisms of Mechanosensitive Piezo Channels. Neuron. 2016;89: 1248-1263.
8. Zhao Q, Zhou H, Chi S, Wang Y, Wang J, Geng J et al. Structure and mechanogating mechanism of the Piezo1 channel. Nature. 2018;554:487-492.
Copyright
© This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/