J Med Discov (2018); 3(1):jmd17057; DOI:10.24262/jmd.3.1.17057; Received September 18th, 2017, Revised January 08th, 2018, Accepted January 22nd, 2018, Published January 28th, 2018.
Application of CRISPR-Cas9 gene editing system: non-viral delivery strategies and improvements
Ni Deng1, Yong-Jun Chen2, 3, *
1College of Medicine, Yangzhou University, Yangzhou 225001, Jiangsu, P. R. China
2People’s Hospital of Hongze District, Huai-An 223100, Jiangsu, P. R. China
3Hongze Hospital of Traditional Chinese Medicine, Huai-An 223100, Jiangsu, P. R. China
* Correspondence: Yong-Jun Chen, Address: People’s Hospital of Hongze District, 102 Dongfeng Rd, Hongze Xian, Huai an City, Jiangsu Province, 223100, P. R. China. Tel: +86 0517-87283427, Fax:+0517-87283416. E-mail: syj3100@126.com
Abstract
Clustered regularly interspaced short palindromic repeats associated protein-9 nuclease (CRISPR-Cas9), known as a heritable part of the prokaryotic adaptive immune system, has been reprocessed for gene editing in mammalian cells. Guide RNA (gRNA) is responsible for recognizing target sequence in genome and following Cas9 protein is applied for cutting two strands of DNA. CRISPR-Cas9 is not the first emerged gene-engineering tool, however, it is versatile, precise and simple over its predecessors such as TALENS and Zinc finger nuclease. As a versatile and powerful gene editing platform, CRISPR-Cas9 gene editing system has been widely applied in biologic research, human medicine, and biotechnology since its discovery. CRISPR-Cas9 gene editing system possesses bright prospects in genetic disorders therapies through revising unwanted mutations caused by disease. Some encouraging results have been achieved by application of CRISPR-Cas9 gene editing system for gene therapy in preliminary research. Despite the promising consequences, some drawbacks still exist in CRISPR-Cas9 gene editing system. Additionally, delivery of CRISPR-Cas9 gene editing system, one of the greatest challenges, remains to be settled, before employing CRISPR-Cas9 system for therapy. In this review, improvements of CRISPR-Cas9 gene editing system will be discussed and non-viral delivery systems for CRISPR-Cas9 gene editing system will be highlighted.
Keywords: CRISPR, CRISPR-Cas9, Delivery systems, Gene editing
Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR)/ CRISPR-associated (Cas), as a heritable and part of the adaptive immune system in archaea and bacteria, is used to defend invasive phages and plasmids. Currently, CRISPR/Cas9 has been successfully reprocessed for gene editing in mammalian cells. Guide RNA (gRNA) is designed to recognize target sequence in genome and followed by Cas9 protein acting as a pair of scissors to cut two strands of DNA. CRISPR-Cas9 gene editing system has triggered a research boom due to its versatile, precise and simple to use. In the past several years, it has been widely applied in biologic research [1], human medicine [2], biotechnology [3] and agriculture [4].
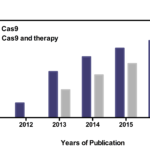
CRISPR structure was found by Yoshizumi Ishino as early as 1987 [5], and the CRISPR was proposed in 2001 [6, 7]. A turning point was appeared in 2005 due to observation that sequences of hyper variable spacers had homology with foreign sequences. Therefore, it is speculated that CRISPR and its related proteins may possess immune memory function, playing a key role against foreign invasion sequences [8]. After that, details of this system were clearly demonstrated because of accelerating pace of development of CRISPR. Zhang, Doudna and Charpentier are three important contributors for development and application of CRISPR-Cas9 gene editing system. Charpentier demonstrates the mechanism of CRISPR-Cas9 system [9]. Furthermore, biochemical features of CRISPR-Cas9 gene editing system were elucidated by Charpentier and Doudna [10]. Zhang adopts the system for gene editing in human cell [11]. CRISPR-Cas9 gene editing system was marked as breakthrough of the year by Science in 2015[12]. The publications about CRISPR-Cas9 are significantly increasing since 2011(Fig 1).
CRISPR-Cas9 is a novel system for gene editing, however, its simplicity, versatile, and precise attracts researchers. On the other hand, some drawbacks were reported in the application of CRISPR-Cas9 gene editing system. Furthermore, the non-viral mediated delivery of CRISPR-Cas9 is a crucial aspect for its application. Therefore, this review aims to discuss the non-viral mediated delivery systems, improvement and drawbacks of CRISPR-Cas9 gene editing system.
Mechanism of CRISPR-Cas9 genome editing system
The CRISPR-Cas system has Class1 and Class 2 [13]. The Class 2 CRISPR-Cas system has one multi-domain protein, which is easier to use than Class 1 CRISPR-Cas system with the large, multi-subunit protein [14]. Type II CRISPR system, also known as CRISPR-Cas9 gene editing system, is the most widely applied among Class 2 family. In type II CRISPR system, CRISPR spacers is responsible for the target sequences, and Cas9 protein is used for acquisition and defense. The action of CRISPR-Cas9 gene editing system has three stages, which are discussed below [14-19].
Adaptation
In this stage, invasion DNA sequences, also known as protospacers, are incorporated. These protospacers are from plasmids or invading viruses, inserting into CRISPR array as new spacers. New spacers are responsible for providing the sequence memory for targeted defenses against invasive sequences.
Expression
In this stage, the CRISPR repeat-spacer array is transcribed into precursor transcript, called pre-crRNA, which is further processed and matured into a crRNAs. The crRNAs have a conserved repeating sequence and a variable spacer sequence complementary to the invasion sequences. The crRNAs recognize large numbers of foreign sequences due to each crRNA responsible for one specific foreign sequence.
Interference
In this stage, crRNAs specifically recognize PAM sequences and Cas9 protein is responsible for cleaving target DNA or RNA. In type II CRISPR-Cas9 system, gRNA is responsible for targeting sequence and following Cas9 protein cuts double strands of DNA and inducing Double Strand Break (DSB), thereby preventing proliferation of foreign gene sequences.
Non-Homologous End Joining (NHEJ) repair pathway and Homology Directed Repair (HDR) pathway
Cas9 protein cuts 3-4 nucleotides upstream of the protospacer adjacent motif (PAM) sequence. Two different pathways called NHEJ repair pathway and HDR pathway, respectively, were used to repair after DSB. The NHEJ repair pathway causes inserts or deletions at the DSB site, further producing frame shifts or premature stop codons. The NHEJ repair pathway disrupts the open reading frame in the targeted sequences. The HDR pathway is used to repair the DSB in the presence of template DNA. The specific template DNA sequences can be inserted due to HDR inducing the copy of repair template to the cleaved target sequence. Gene replacement or knock in has lower efficiency in comparison to gene knock out by using CRISPR-Cas9 gene editing system, as initiation of HDR repaired pathway has lower possibility than NHEJ repaired pathway after DSB.
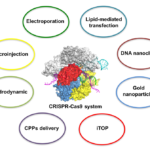
Delivery systems for CRISPR-Cas9 gene editing system
In early stage for delivery of CRISPR-Cas9 gene editing system, non-viral delivery systems such as electroporation [20], hydrodynamic delivery [21-24], and lipid-based nanoparticles [25] are major methods. There are several advantages for non-viral delivery systems. First, non-viral delivery systems have higher safety than viral mediated delivery systems. Second, non-viral delivery systems have no limitation for length of DNA sequences, which is a big challenge for viral-mediated delivery systems. Third, non-viral delivery systems are easy to produce at relatively lower cost than that of viral-mediated delivery system [26-29]. In this section, current non-viral delivery systems used to deliver CRISPR-Cas9 gene editing system are discussed.
Electroporation
Electroporation is an approach used to deliver macromolecules into mammalian cells such as protein and nucleotides including DNA and RNA [30]. In the process of electroporation, permeability is temporarily increasing in cell membrane due to electrical field, which allows macromolecules such as nucleotides and proteins to cross cytoplasm membrane or nuclear membrane [31]. Electroporation is desirable for plasmid-based CRISPR-Cas9 gene editing system systems such as pX260 and pX330 system, Cas9 mRNA and gRNA, Cas9 protein combined gRNA (RNP) and templates DNA. For plasmid-based CRISPR-Cas9 gene editing system, electroporation has been achieved many encouraging results in vertebrate organogenesis field. zebrafish fin and axolotl have been successfully regenerated by electroporation of plasmids encoding Cas9 protein and gRNA [32-35]. Moreover, electroporation has also been used for delivery of plasmids encoding Cas9 protein and gRNA into embryonic stem cell, T lymphocytes and different cancer cell lines [36-41]. Unfortunately, only around 0.01% of target cells have been achieved by using electroporation of plasmid-based CRISPR-Cas9 gene editing system. Plus, electroporation results in cell damage and high percentage of cell death. Electroporation is desirable for delivery of Cas9 mRNA and gRNA. Qin and colleagues deliver Cas9 mRNA and gRNA into mouse zygotes using electroporation, achieving ideal genotype [42]. Furthermore, electroporation is also desirable for delivery of RNP. Fei and colleagues demonstrate that RNP has higher efficiency than plasmid-based CRISPR-Cas9 gene editing system in the cells by using electroporation [43]. Furthermore, electroporation can be used to deliver RNPs in different cell lines including embryonic stem cells, fibroblasts [44], and human T lymphocytes [45]. Liang and colleagues make a comparison among plasmid based-, mRNA based- and RNPs based-systems. The results demonstrate that RNP has highest gene-editing efficiency than other two systems by using electroporation. For example, electroporation of RNPs has over 94% editing efficiency in Jurkat T cells and 87% editing efficiency in pluripotent stem cells. For plasmid-based system, only 63% editing efficiency in Jurkat T cells and 20% editing efficiency in pluripotent stem cells are achieved. And also, electroporation of mRNA-based system achieves low efficiency with 32% editing efficiency in pluripotent stem cells and 42% editing efficiency in Jurkat T cells [46].
Lipid-mediated transfection
Lipid nanoparticle as a commonly used strategy for delivery of nucleic acids, has entered Phase I clinical trials for RNAi therapy with encouraging results [47, 48]. The positively charged lipids and negatively charged nucleic acids form complexes by charge-charge interaction. Cells uptake the complexes by endocytosis or macro-pinocytosis. By now, lipid-mediated delivery of CRISPR-Cas9 gene editing system has been used in construction of cell models [11, 40] and even in establishment of animal models [49, 50].
As mentioned, RNPs has higher gene editing efficiency than plasmid-based CRISPR-Cas9 gene editing system. Zuris and colleagues demonstrate that cationic lipid-mediated RNP delivery achieves over 80% modification in comparison to plasmid-based system in mammalian cells. This study also determines that lipid-mediated RNP delivery achieves around 20 % gene editing efficiency in hair cells in vivo [25]. Furthermore, bio-reducible lipids were designed to deliver RNP. Bio-reducible lipid degrades in the cytosol, and then enhances the endosome escape of RNP. By using this method, over 70 % gene editing efficiency were achieved [51]. The results demonstrate that lipid-mediated RNP delivery is an ideal method to target certain cells such as HEK293FT and N2A. Lipid-mediated transfection also can be used for delivery of plasmid-based CRISPR-Cas9 gene editing system or Cas9 mRNA/gRNA for some cell lines such as HEK293FT, U2OS, mouse ESCs and N2A [46]. Therefore, lipid-mediated delivery of CRISPR-Cas9 gene editing systems is a desirable method for gene editing in vitro and in vivo.
Hydrodynamic delivery
Hydrodynamic delivery is a safe and convenient method for delivery of DNA in vivo in comparison to viral delivery system. Mechanical force is generated with rapidly injection of DNA solution with large volume in blood vessel, generating pores on cell membrane of endothelial cells and enable DNA to enter cells [52-54]. Recently, several researches report that plasmid-based CRISPER-Cas9 gene editing system can be successfully applied and generates genetic corrections or mutations using hydrodynamic injection [22, 23, 55, 56]. Yin and colleagues construct mice phenotype using hydrodynamic tail injection of plasmid-based system. pX330 system was used to combine with corrected DNA template of Fah. Hydrodynamic tail injection was used to delivery pX330 system and template DNA, leading to expression of Fah protein in ~1/250 liver cells [57]. Similarly, Hydrodynamic tail injection was also used to delivery pX330 system targeting Pten sequences, achieving around 2.6 % of sequence with mutations in liver [58]. Therefore, hydrodynamic injection of plasmid-based system is a good method for construction of hepatoma models. On the other hand, hydrodynamic injection has several drawbacks such as inducing the increase of blood pressure, causing liver expansion and dysfunction of temporary cardiac [53]. Besides, application of hydrodynamic injection in the large animals is still not clear, and hydrodynamic injection is hard to use in clinical implementation.
Microinjection
Microinjection is desirable for delivery of plasmid-based or mRNA-based systems [59-62]. Injection of plasmid-based system into pronucleus is an easiest way for achieving ideal genotype [63-65]. some studies demonstrate that microinjection of plasmid-based or mRNA-based systems can be applied in vivo targeted genetic modification in rabbits [66], zebrafish [67], Ciona intestinalis [68], worms[69] and Aedes aegypti [70]. In addition, microinjection of the plasmid-based CRISPR-Cas9 gene-editing system is also useful for multi-targeting [66].
Nakagawa and colleagues compare gene editing efficiency based on plasmid-based systems, 1) Cas9 nuclease with one gRNA, 2) Cas9 nickase with two gRNAs, 3) FokI-dCas9 with two gRNAs via using microinjection. The results indicate that Cas9 nickase results in lowest mutation rates and the highest birth rates. By the contrary, Cas9 nuclease leads to lowest birth rates and the highest mutation rates [71]. On the other hand, microinjection of plasmid-based system induces unwanted integration in the host chromosomes. Alternatively, microinjection of mRNA-based system into pronucleus has high efficiency, avoiding unwanted interstation. In addition, the delivery site is very important for microinjection. For instance, Horii and colleagues demonstrate that microinjection of mRNA based system into pronucleus achieve higher gene editing efficiency than microinjection of mRNA-based system into cytoplasm or microinjection of plasmid-based system in pronucleus [72]. The results demonstrate microinjection as an effective physical method for delivery of plasmid-based or mRNA-based systems. However, some disadvantages of microinjection should not be ignored. First, microinjection leads to cell damage. Second, it requires complicated manual skills. Third, numbers of cells are limited for microinjection because only one cell can be targeted at each time.
Induced transduction by osmocytosis and propanebetaine (iTOP)
iTOP, as a novel emerged method for delivery of CRISPR-Cas9 gene editing system, is an active uptake process. A buffer containing sodium chloride-mediate hyperosmolality combined with propanbetaine, a transduction compound, stimulates cell macro-pinocytotic uptake and further delivered RNP. iTOP transduction has been successfully used for delivery of Cas9 protein and gRNA, separately. Besides, it also has been used for delivery of RNP into a variety of primary cells. According to this method, Cas9 protein and gRNA were co-transduced in human embryonic stem cells with 10% and 26% gene-editing frequency after one and two rounds of transduction, respectively [26]. In comparison to other delivery systems such as electroporation and lipid nanoparticles, iTOP shows low gene editing efficiency [25, 26, 73-75]. Another drawback of iTOP is that Cas9 protein is hard to soluble in low salt solution, which is not desirable for application in in vivo study.
Cell-penetrating peptides (CPPs)
CPPs are an emerged delivery system for delivery CRISPR-Cas9 gene editing system. Briefly, CPPs were covalently conjugated with Cas9 protein and gRNA complexes with another peptide. Those two carriers were co-delivered in the cells, further generating mutation ranging from 2.3% to 16% among different cell lines including HEK293T, HeLa, dermal fibroblasts, and embryonic cells [74, 75]. However, RNPs and CPP are failed to complex due to gRNA neutralization positive charged CPP and the cellular entry of RNPs-CPP complex is blocked. Based on this result, we speculate the application of CPP-mediated CRISPR-Cas9 gene editing system delivery is less likely in vivo. Besides, since gRNA-CPP is highly cationic, so it is not favored for circulation kinetics or bio-distribution.
DNA nanoclews
Yarn-like DNA nanoclews are a novel emerged method for delivery of RNP [76]. DNA nanoclews are made of amplification of rolling circle of DNA. Due to structural homology to the gRNA, RNP can complex into the DNA cage. PEI was added to the complex, enhancing cell uptake and endosome escaping of RNP. Delivery of RNP using DNA nanoclews achieves 28% gene editing efficiency in U2OS-EGFP cells. Also, DNA nanoclews mediated RNP delivery leads to around 25% EGFP disruption in tumor via intra-tumor injection [77]. However, due to high toxicity induced by PEI and immunogenicity, it is less likely for using this system in human body.
Gold nanoparticles
Gold nanoparticles is used to co-assemble Cas9 protein labeled a glutamate peptide tag with gRNA into nano-assemblies. The cellular uptake of gold nanoparticle-RNP dependents on cholesterol-dependent membrane fusion other than cellular endocytosis [78]. Over 30% gene editing efficiency can be achieved by using gold nanoparticles mediated delivery system in different cell lines [78], providing an alternative delivery method for CRISPR-Cas9 gene editing delivery. However, further studies are still necessary to evaluate the efficiency of system in human body.
Fig 1. Publications related to “Cas9” and “Cas9 and therapy” for each year since 2011. For 2017, the numbers of publications are basing to the publications from January 1th to December 17th.
Improvement of CRISPR-Cas9 gene editing system
CRISPR-Cas9 gene editing system possesses bright prospects in therapy of genetic disorders via directly revising unwanted gene mutations. Until now, many advances in the development of specificity, repair and delivery strategies for the CRISPR-Cas9 gene editing system have been achieved. Specifically, some aspects are under rapidly development including improvement of Cas9 nuclease specificity, optimization of gRNA, and detection methods with higher efficiency for off-target. These improvements increase efficiencies of delivery and gene editing of system. Herein, we briefly summarize recent improvements for CRISPR-Cas9 gene editing system.
With structure analysis of Cas9 nuclease, engineered Cas9 nucleases with high gene editing efficiency are rapidly developed. Cas9 nuclease contains two catalytic domains called RuvC and HNH, respectively. Once one point mutation is introduced in Cas9 catalytic domains, Cas9 protein, also called Cas9 nickase, only cleave one strand DNA complementary to the gRNA instead of two strands of DNA. Therefore, Cas9 nickase reduces off-target effect (50- to 1500-fold) and induces high efficiency of HDR comparing to Cas9 nuclease [11, 79]. In addition, a nuclease-inactive Cas9 protein, also called dCas9 can be generated with mutations of two Cas9 catalytic domains [80]. Furthermore, the dimerization-dependent system produced by fusion dCas9 protein with FokI nuclease domain has a higher specificity (over 140-fold) than normal Cas9 protein[81]. A similar system is fusion Cas9 nuclease with programmable DNA-binding domain, thereby reducing inherent DNA-binding affinity of Cas9 nuclease. This system also dramatically reduces off-target effects [82].
A novel structure-guided engineering strategy based on Cas9 protein without adding extra components has been developed. SpCas9 and SaCas9 with high specificity were produced by mutations non-DNA binding sites of Cas9 protein domain, improving on-target effects [83]. Similarly, Kleinstiver and colleagues develop a high-fidelity variant SpCas9 protein with over 70% efficiency. It contains four alanine substitutions, enhancing interaction effects of Cas9 protein with target DNA [84]. On the other hand, with emerging of these Cas9 variants, detection methods for off-target effects with high accuracy should be developed. And also, the current delivery systems should take advantage of Cas9 variants, which may achieve higher gene editing efficiency in comparison to wide-type Cas9 protein.
Modified gRNA is another available component for enhancing specificity of CRISPR-Cas9 gene editing system. In general, RNA duplex is composed of CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA), which are responsible for targeting and cleavage, respectively. A synthetic single strand gRNA (sgRNA) is more advanced, containing crRNA and tracrRNA in single strand, mediating Cas9 protein targeting and cleaving double strands of DNA. By structure analysis of gRNA, Briner and colleagues discovered six features of gRNA, which are responsible for CRISPR-Cas9 targeting, indicating that modifications of these six features may increase the on-target effects [85]. Another method to improve editing efficiency of Cas9 protein is extension of gRNA. Five bp extension of the gRNA duplex improves on-target efficiency of dCas 9 in one experiment [86]. Meanwhile, the experiment demonstrate that knockout efficiency of Cas9 will reach a peak with 5 bp extension of the gRNA duplex [87]. Hsu and colleagues also determine that extension the gRNA duplex to 10 bp without improving knockout efficiency [88].
Length of guide sequence is a crucial factor, determine specificity of CRISPR-Cas9 gene editing system. The results demonstrate that truncated guide sequence-mediated CRISPR-Cas9 gene editing system possesses highly precise on-target effects. Fu and colleagues demonstrate if the length of guide sequence is less than 20 bp (17 bp or 19 bp), it significantly reduces mutations at some off-target sites without reducing on-target effects. However, it should be notice that guide sequence with 15 nucleotides is too short to have guidance effects [89]. Furthermore, although shorter guide sequences mediated gene editing has lower off-target activity, but more off-target sites are generated[90]. Additionally, even combined these shorter guide sequences with high fidelity SpCas9, low on-target efficiency were still being detected [84]. Therefore, the combination of optimized gRNA with engineered Cas9 protein should be further investigated.
Chemical modification of gRNA also can improve efficiency of CRISPR-Cas9 gene editing system. Modification of gRNA at 3’ and 5’ ends with 2’ -O-methyl 3’ thioPACE, 2’-O-methyl 3’phosphorothioate or 2’-O-methyl significantly improves gene-editing efficiency of CRISPR-Cas9 gene editing system in a variety of cell lines [91]. Increased off-target effect is one major drawback of modification gRNA, indicating that modification of gRNA should be further explored to reduce off-target effects.
Perspective and conclusion
Many improvements for CRISPR-Cas9 gene editing system are under development, off-target effect is still a major limitation as off-target may induce genome instability, gene functional disruptions and epigenetic alterations. In addition, many potential off-target sites may exist in large genomes, such as in mammals, because relatively small gRNA targeting DNA sequences only through 20 bp binding. Another possibility is the occurrence of non-Watson–Crick base pairing, leading to more off-target activities of CRISPR-Cas9 gene editing system [92]. According to two studies performed by Cradick [93] and Fu [89], at least 1/3 large insertions and 23% small insertions at off-target sites based on plasmid-based CRISPR-Cas9 gene editing system. It is known that unwanted insertions at off-target sites are difficult to detect and it is more problematic than those insertions at on-target sites because foreign sequences can induce host immune responses. Therefore, to better take advantage of CRISPR-Cas9 gene editing system, some strategies such as optimization of Cas9 protein, rational design of gRNA(s), and selection of targeting site should be developed before application of CRISPR-Cas9 gene editing system.
In past few years, the CRISPR-Cas9 gene editing system has been widely used for gene editing and achieved encouraging results in biological fields. However, delivery of CRISPR-Cas9 gene editing system remains a big challenge. Therefore, development of more advanced delivery systems is very important for application of CRISPR-Cas9 gene editing system in vivo study. Currently, some delivery systems including non-viral-mediated and viral-mediated systems have been developed for delivery of CRISPR-Cas9 gene editing system. Based on these delivery systems, CRISPR-Cas9 gene editing system-based therapeutics have been achieved some promising results. In the future, to apply CRISPR-Cas9 gene editing system in human body for therapeutic purpose, more advanced delivery systems with high efficiency and low immunogenicity should be further investigated.
Conflict of interest
None
Acknowledgments
None
References
1. Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720-3.
2. Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, et al. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27-30.
3. Sampson TR, Weiss DS. Exploiting CRISPR/Cas systems for biotechnology. Bioessays. 2014;36:34-8.
4. Khatodia S, Bhatotia K, Passricha N, Khurana SM, Tuteja N. The CRISPR/Cas Genome-Editing Tool: Application in Improvement of Crops. Front Plant Sci. 2016;7:506.
5. Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429-33.
6. Jansen R, van Embden JDA, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Molecular Microbiology. 2002;43:1565-75.
7. Jansen R, van Embden JD, Gaastra W, Schouls LM. Identification of a novel family of sequence repeats among prokaryotes. Omics. 2002;6:23-33.
8. Mojica FJM, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174-82.
9. Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao YJ, Pirzada ZA, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602-+.
10. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816-21.
11. Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819-23.
12. Travis J. GENETIC ENGINEERING. Germline editing dominates DNA summit. Science. 2015;350:1299-300.
13. Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722-36.
14. van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479-92.
15. Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67-71.
16. Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467-77.
17. Gasiunas G, Sinkunas T, Siksnys V. Molecular mechanisms of CRISPR-mediated microbial immunity. Cell Mol Life Sci. 2014;71:449-65.
18. Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096.
19. Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release. 2017;266:17-26.
20. Qin WN, Dion SL, Kutny PM, Zhang YF, Cheng AW, Jillette NL, et al. Efficient CRISPR/Cas9-Mediated Genome Editing in Mice by Zygote Electroporation of Nuclease. Genetics. 2015;200:423-+.
21. Gori JL, Hsu PD, Maeder ML, Shen S, Welstead GG, Bumcrot D. Delivery and Specificity of CRISPR/Cas9 Genome Editing Technologies for Human Gene Therapy. Human Gene Therapy. 2015;26:443-51.
22. Yin H, Xue W, Chen SD, Bogorad RL, Benedetti E, Grompe M, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype (vol 32, pg 551, 2014). Nat Biotechnol. 2014;32:952-.
23. Xue W, Chen SD, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380-+.
24. Liu C, Zhang W, Yang H, Sun W, Gong X, Zhao J, et al. A water-soluble inclusion complex of pedunculoside with the polymer beta-cyclodextrin: a novel anti-inflammation agent with low toxicity. PLoS One. 2014;9:e101761.
25. Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33:73-80.
26. D’Astolfo DS, Pagliero RJ, Pras A, Karthaus WR, Clevers H, Prasad V, et al. Efficient intracellular delivery of native proteins. Cell. 2015;161:674-90.
27. Li L, Natarajan P, Allen C, Peshwa MV. Cgmp-Compliant, Clinical Scale, Non-Viral Platform for Efficient Gene Editing Using Crispr/Cas9. Cytotherapy. 2014;16:S37-S.
28. Katz MG, Fargnoli AS, Williams RD, Bridges CR. Gene Therapy Delivery Systems for Enhancing Viral and Nonviral Vectors for Cardiac Diseases: Current Concepts and Future Applications. Human Gene Therapy. 2013;24:914-27.
29. Liu C, Zhao J, Liu Y, Huang Y, Shen Y, Wang J, et al. A novel pentacyclic triterpenoid, Ilexgenin A, shows reduction of atherosclerosis in apolipoprotein E deficient mice. Int Immunopharmacol. 2016;40:115-24.
30. Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, et al. Gene Editing of CCR5 in Autologous CD4 T Cells of Persons Infected with HIV. New England Journal of Medicine. 2014;370:901-10.
31. Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823-6.
32. Thummel R, Bai S, Sarras MP, Song PZ, Zhang XM, Hyde DR, et al. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Biol. 2006;295:356-.
33. Nakamura H, Katahira T, Sato T, Watanabe Y, Funahashi J. Gain- and loss-of-function in chick embryos by electroporation. Mech Develop. 2004;121:1137-43.
34. Fei JF, Haffner C, Huttner WB. 3 ‘ UTR-Dependent, miR-92-Mediated Restriction of Tis21 Expression Maintains Asymmetric Neural Stem Cell Division to Ensure Proper Neocortex Size. Cell Reports. 2014;7:398-411.
35. Ni L, Wang J, Liu C, Fan J, Sun Y, Zhou Z, et al. An asymmetric binuclear zinc(ii) complex with mixed iminodiacetate and phenanthroline ligands: synthesis, characterization, structural conversion and anticancer properties. Inorg Chem Front. 2016.
36. Yang H, Wang HY, Shivalila CS, Cheng AW, Shi LY, Jaenisch R. One-Step Generation of Mice Carrying Reporter and Conditional Alleles by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;154:1370-9.
37. Mandal PK, Ferreira LMR, Collins R, Meissner TB, Boutwell CL, Friesen M, et al. Efficient Ablation of Genes in Human Hematopoietic Stem and Effector Cells using CRISPR/Cas9. Cell Stem Cell. 2014;15:643-52.
38. Hou ZG, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:15644-9.
39. Ding QR, Regan SN, Xia YL, Oostrom LA, Cowan CA, Musunuru K. Enhanced Efficiency of Human Pluripotent Stem Cell Genome Editing through Replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12:393-4.
40. Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833-+.
41. Sun W, Liu C, Zhang Y, Qiu X, Zhang L, Zhao H, et al. Ilexgenin A, a novel pentacyclic triterpenoid extracted from Aquifoliaceae shows reduction of LPS-induced peritonitis in mice. Eur J Pharmacol. 2017.
42. Qin W, Dion SL, Kutny PM, Zhang Y, Cheng AW, Jillette NL, et al. Efficient CRISPR/Cas9-Mediated Genome Editing in Mice by Zygote Electroporation of Nuclease. Genetics. 2015;200:423-30.
43. Fei J-F, Knapp D, Schuez M, Murawala P, Zou Y, Singh SP, et al. Tissue-and time-directed electroporation of CAS9 protein–gRNA complexes in vivo yields efficient multigene knockout for studying gene function in regeneration. npj Regenerative Medicine. 2016;1:16002.
44. Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Research. 2014;24:1012-9.
45. Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, Gate RE, et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. P Natl Acad Sci USA. 2015;112:10437-42.
46. Liang XQ, Potter J, Kumar S, Zou YF, Quintanilla R, Sridharan M, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. Journal of Biotechnology. 2015;208:44-53.
47. Fitzgerald K, Frank-Kamenetsky M, Shulga-Morskaya S, Liebow A, Bettencourt BR, Sutherland JE, et al. Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: a randomised, single-blind, placebo-controlled, phase 1 trial. Lancet. 2014;383:60-8.
48. Coelho T, Adams D, Silva A, Lozeron P, Hawkins PN, Mant T, et al. Safety and Efficacy of RNAi Therapy for Transthyretin Amyloidosis. New Engl J Med. 2013;369:819-29.
49. Platt RJ, Chen SD, Zhou Y, Yim MJ, Swiech L, Kempton HR, et al. CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell. 2014;159:440-55.
50. Raghavan A, Wang X, Rogov P, Wang L, Zhang X, Mikkelsen TS, et al. High-throughput screening and CRISPR-Cas9 modeling of causal lipid-associated expression quantitative trait locus variants. bioRxiv. 2016:056820.
51. Wang M, Zuris JA, Meng FT, Rees H, Sun S, Deng P, et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. P Natl Acad Sci USA. 2016;113:2868-73.
52. Suda T, Liu D. Hydrodynamic delivery. Adv Genet. 2015;89:89-111.
53. Suda T, Liu D. Hydrodynamic gene delivery: its principles and applications. Mol Ther. 2007;15:2063-9.
54. Yang H, Liu C, Zhang YQ, Ge LT, Chen J, Jia XQ, et al. Ilexgenin A induces B16-F10 melanoma cell G1/S arrest in vitro and reduces tumor growth in vivo. Int Immunopharmacol. 2015;24:423-31.
55. Zhen S, Hua L, Liu YH, Gao LC, Fu J, Wan DY, et al. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther. 2015;22:404-12.
56. Yang H, Wang J, Fan JH, Zhang YQ, Zhao JX, Dai XJ, et al. Ilexgenin A exerts anti-inflammation and anti-angiogenesis effects through inhibition of STAT3 and PI3K pathways and exhibits synergistic effects with Sorafenib on hepatoma growth. Toxicol Appl Pharmacol. 2017;315:90-101.
57. Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551-3.
58. Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380-4.
59. Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell research. 2013;23:465-72.
60. Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836-43.
61. Crispo M, Mulet AP, Tesson L, Barrera N, Cuadro F, dos Santos-Neto PC, et al. Efficient Generation of Myostatin Knock-Out Sheep Using CRISPR/Cas9 Technology and Microinjection into Zygotes. Plos One. 2015;10.
62. Zhang W, Gong X, Liu C, Piao Y, Sun Y, Diao G. Water-soluble inclusion complex of fullerene with γ-cyclodextrin polymer for photodynamic therapy. Journal of Materials Chemistry B. 2014;2:5107.
63. Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, Ikawa M. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep-Uk. 2013;3.
64. Mashiko D, Young SAM, Muto M, Kato H, Nozawa K, Ogawa M, et al. Feasibility for a large scale mouse mutagenesis by injecting CRISPR/Cas plasmid into zygotes. Development Growth & Differentiation. 2014;56:122-9.
65. Zhang YQ, Yang H, Sun WD, Wang J, Zhang BY, Shen YJ, et al. Ethanol extract of Ilex hainanensis Merr. exhibits anti-melanoma activity by induction of G1/S cell-cycle arrest and apoptosis. Chin J Integr Med. 2017.
66. Yan Q, Zhang Q, Yang H, Zou Q, Tang C, Fan N, et al. Generation of multi-gene knockout rabbits using the Cas9/gRNA system. Cell Regeneration. 2014;3:12.
67. Hruscha A, Schmid B. Generation of Zebrafish Models by CRISPR/Cas9 Genome Editing. Neuronal Cell Death: Methods and Protocols. 2015;1254:341-50.
68. Sasaki H, Yoshida K, Hozumi A, Sasakura Y. CRISPR/Cas9‐mediated gene knockout in the ascidian Ciona intestinalis. Development, growth & differentiation. 2014;56:499-510.
69. Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nature Methods. 2013;10:741-+.
70. Basu S, Aryan A, Overcash JM, Samuel GH, Anderson MAE, Dahlem TJ, et al. Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. P Natl Acad Sci USA. 2015;112:4038-43.
71. Nakagawa Y, Sakuma T, Sakamoto T, Ohmuraya M, Nakagata N, Yamamoto T. Production of knockout mice by DNA microinjection of various CRISPR/Cas9 vectors into freeze-thawed fertilized oocytes. Bmc Biotechnol. 2015;15.
72. Horii T, Arai Y, Yamazaki M, Morita S, Kimura M, Itoh M, et al. Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Sci Rep-Uk. 2014;4.
73. Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012-9.
74. Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24:1020-7.
75. Suresh B, Ramakrishna S, Kim H. Cell-Penetrating Peptide-Mediated Delivery of Cas9 Protein and Guide RNA for Genome Editing. Methods Mol Biol. 2017;1507:81-94.
76. Sun WJ, Ji WY, Hall JM, Hu QY, Wang C, Beisel CL, et al. Self-Assembled DNA Nanoclews for the Efficient Delivery of CRISPR-Cas9 for Genome Editing. Angewandte Chemie-International Edition. 2015;54:12029-33.
77. Sun W, Ji W, Hall JM, Hu Q, Wang C, Beisel CL, et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew Chem Int Ed Engl. 2015;54:12029-33.
78. Mout R, Ray M, Yesilbag Tonga G, Lee YW, Tay T, Sasaki K, et al. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano. 2017.
79. Friedland AE, Baral R, Singhal P, Loveluck K, Shen S, Sanchez M, et al. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biology. 2015;16.
80. Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, et al. Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell. 2014;156:935-49.
81. Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577-82.
82. Bolukbasi MF, Gupta A, Oikemus S, Derr AG, Garber M, Brodsky MH, et al. DNA-binding-domain fusions enhance the targeting range and precision of Cas9. Nat Methods. 2015;12:1150-6.
83. Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84-8.
84. Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng ZL, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490-+.
85. Briner AE, Donohoue PD, Gomaa AA, Selle K, Slorach EM, Nye CH, et al. Guide RNA Functional Modules Direct Cas9 Activity and Orthogonality. Molecular Cell. 2014;56:333-9.
86. Chen BH, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System (vol 155, pg 1479, 2013). Cell. 2014;156:373-.
87. Dang Y, Jia GX, Choi J, Ma HM, Anaya E, Ye CT, et al. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biology. 2015;16.
88. Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827-+.
89. Fu YF, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279-84.
90. Slaymaker IM, Gao LY, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84-8.
91. Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015;33:985-U232.
92. Jiang WY, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233-9.
93. Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584-92.
Copyright
© This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/