J Med Discov (2017); 2(3): jmd17035; DOI: 10.24262/jmd.2.3.17035; Received August 1st, 2017, Revised September 5th, 2017, Accepted September 17th, 2017, Published September 20th, 2017
Detection of genetic variations from next-generation sequencing data
Ying Wang1, Xiaopeng zhu2, and Jun Ding3*
1NSABP Foundation, Inc, Pittsburgh, PA, 15212, USA
2, 3Computational Biology Department, School of Computer Science, Carnegie Mellon University, Pittsburgh, PA 15213, USA.
* Correspondence: Jun Ding, Computational Biology Department, School of Computer Science, Carnegie Mellon University, Pittsburgh, PA 15213, USA. jund@cs.cmu.edu.
Abstract
Genetic variation describes the differences between individual genomes and next generation sequencing (NGS) is becoming more and more important for studying the association between the genetic variations and disease-related phenotypes. In this review, we discussed the general pipeline for detecting genetic variation from high-throughput NGS reads. For each component of the pipeline, we compared several widely used tools, which could be very helpful for users to choose appropriate tools and parameters to call and analyze the genetic variants under their specific circumstances.
Keywords: Genetic variation, next generation sequencing.
Introduction
With the advancement of high-throughput sequencing technique, the cost of genome sequencing is decreasing rapidly in the past decade, leading to the great achievements in personal genome sequencing projects such as 1000 genome project [1] and the personal genome project [2]. The increasing availability of the genome sequences provides a great opportunity to study the genetic variations between different entities.
Genetic variation describes the differences between our genomes. There are two important types of genetic variations: ‘Single nucleotide polymorphism’ (SNP) and ‘Structure variation’. SNP is the difference in a single nucleotide between members of one species, which is the most common type of the genetic variation, estimated to account for 90% of all variants [3]. Structural variation is the variation in the structure of the genome, such as copy-number variation, deletions, inversions, insertions and duplications. While some genetic variation is harmless, many of the genetic variations are playing important roles in diverse types of diseases such as Crohn’s disease [4]. Therefore, detecting and studying those genetic variations is of vital importance to understand the underlying mechanism, which can help us to diagnose or even cure the corresponding diseases. Already many Mendelian disease studies have employed NGS techniques to identify causal genes based on patient-specific variants [5, 6, 7]. As Mendelian disease related variations rarely occur among the healthy genome, the interpretation of patient-specific variations is relatively simple. However, this would potential raise the false discoveries due to the errors in sequencing and false variation detection methods. Therefore, the accurate variation detection method is very critical for the success of the clinical genomic based on NGS techniques.
There are lots of existing pipelines to identify genetic variants. Typically, those pipelines are composed of two major components: Read aligner and Variant caller. The names are quite self-explanatory. Read aligner is used to align reads to the reference genome while the variant caller is used to call the variants based on the mapped reads. In the remaining sections of this work, we will discuss several widely used pipelines for detecting genetic variants and how to study the association between the genetic variants and the phenotype of interest such as certain disease.
Genetic variants detecting pipeline
NGS read aligners
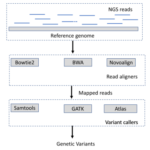
As described in Figure 1, we need first to map the raw NGS reads to the reference genome before calling any potential genetic variants. The mapping accuracy is very important in variation detection. If the reads are aligned
Figure 1: Genetic variants detecting pipeline
incorrectly, the following SNP detecting would be also problematic. Therefore, choosing the read aligner is very important in detecting genetic variations. Among the many read aligners, Bowtie2 [8], BWA [9] and Novoalign ( novocraft.com) are the most popular ones. We will discuss the characteristics of those read aligners so that users may have an idea of how to choose an appropriate read aligner under different scenarios. These popular read aligners typically fall into categories: hash-table indexed or FM-indexed. The idea of hash table indexing can be traced back to BLAST [10]. This hash table based indexing essentially follow the same seed-and-extend procedure as BLAST. First, keeps the positions of k-mer query subsequence as keys. Second, searches for the exact match of the keys, named as seeds, in reference sequences. Third, extends and joins the seeds without gaps and then refines them by a Smith-Waterman alignment [11]. The basic BLAST algorithm has been improved for the alignment of different types. This type of methods typically consumes less space as it builds the index based on the position of sequences instead of the sequences themselves. The FM-indexing is basically based on Burrows-Wheeler transform(BWT) [9]. When a string is transformed by BWT, the order of the characters is permuted. If there are several sub-strings that occurred often in the original string, the transformed string will have several places where a single character is repeated multiple times in a row. BWT transforms the original string into a more compressed format where the same characters are placed side by side as a group rather than in a scattered way and such transformation is reversible. For the 3 aligners we discussed, BWA and Bowtie2 are based on FM-index while Novoalign adopts the hash table indexing as described in Table 1.
Another difference between those aligners is the way to find the inexact matches. As most aligners allow a certain number of mismatches, finding inexact matches efficiently is very important for read aligners. Bowtie utilizes a backtracking strategy to perform a depth-first search through the entire space, which stops until the first alignment that satisfies specific criterion is found. BWA is using a similar backtracking strategy as Bowtie. However, the search in BWA is bounded by the lower limit of number of mismatches in the reads. With a better estimation of the limit, BMA can search in a much smaller space and thus can be much more efficient. Novoalign first finds candidate alignment positions from the reference genome for each read and calculates the alignment score, based on base qualities, the existence of gap and ambiguous codes(Ns). Because of this alignment score based strategy, users are unable to set up allowed mismatches for Novoalign, but they can specify an alignment score instead. Table 1 presents a comparison between different read aligners as presented in [12]. BWT-based aligners (e.g. Bowtie, BWA) are fast, memory-efficient and particularly useful for aligning repetitive reads, however, they tend to be less sensitive than the state-of-art hash-based algorithms (e.g. Novoalign).
The reads mapping is a tradeoff between accuracy and read depth. Using a stringent alignment cutoff (e.g. smaller mismatches or larger alignment score) always lead to fewer mapped reads while using loose cutoff leads to worse alignment accuracy. The optimal choices of alignment cutoffs might differ in different organisms. For example, the populations of fruit-fly are generally more variable than human populations, using alignment cutoff optimized for human sequence analysis might lead to severe loss of mapped reads in fruit-fly. This, in turn, might lead to potential biases for all downstream analysis. On the other hand, using alignment cutoff good for fruit-fly might be leading to a huge amount of incorrectly mapped reads in human. Therefore, it’s very important to choose the right alignment parameters for the mapped reads. Users might need empirical experiences from previous studies to choose ‘proper’ parameters for each of those aligners in different species.
Genetic variants caller
After we got the mapped reads using the read aligners (normally in SAM format or BAM format), we will need to call genetic variants afterwards. Variants calling typically is composed of two components: genotype assignment and variant identification. Early probabilistic methods such as Mapping and Assembly with Quality (MAQ) [13] and SOAPsnp [14],used fixed prior values for heterozygote probabilities and nucleotide-read error probabilities. Multiple sample genotype calling, using EM algorithm to estimate the model parameters, was used by seqEM [15].
The most widely used variant callers include SAMtools [16], GATK [17], Atlas [18]. Although SAMtools and GTAK are both using a Bayesian approach to call the variants, they produce slightly different variants. There are some major differences between GATK and SAMtools, two of the most widely used variant callers. First, GATK drops reads with low mapping quality, but SAMtools uses all reads by default. Second, the model behind SAMtools and GATK is very similar. GATK later augmented the model to work with multi-allelic cases while SAMtools did not. Third, Samtools utilized hand-tune filters while GATK learns filters from data. GATK’s approach is more convenient and powerful especially when we have enough data to train the model. GATK is clearly more complicated and tends to have better variant calling results. As described in [19], the detected variants from GATK were compared with those from SAMtools from the same 30 subjects. For these comparisons, they used the UnifiedGenotyper algorithm in GATK and mpileup in SAMtools. They observed a true-positive rate of 95% for GATK and 70% for SAMtools. Although having relatively worse performance than GATK, SAMTools remains a useful tool for many tasks. As it limits the total read depth to 8000, it is more suitable to evaluate whole genome sequencing data at moderate coverage rather than target candidate gene sequencing, which generally contain a large portion of sites with higher coverage. Besides, the single-sampleSNP detection accuracy is similar to GATK. Compared with SAMtools and GATK, Atlas is based on total different model (logistic regression), which does not utilize the traditional probabilistic model to calculate likelihood. Besides, it calls SNPs and indels using separate programs. As discussed in [20], Atlas did not show consistent advantages over the other methods especially GATK.
| Bowtie2 | BWA | Novoalign | |
| Availability | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | http://bio-bwa.sourceforge.net/ | http://www.novocraft.com/ |
| Indexing | FM-index | FM-index | Hash table |
| Inexact match | Back-tracking | Back-tracking | Alignment score |
| mismatch allowed | 0-3 max in read | up to 6 | up to 8 |
| Alignments reported per read | up to any | up to any | random/all/none |
| Gap alignment | unavailable | available | available |
| Best alignment | minimal number of mismatch | minimal number of mismatch | highest alignment score |
Table 1: read aligners comparison
|
Caller Availability |
SAMTools |
GATK |
Atlas |
| Code | C | Java | Ruby |
| Model | HMM,MAQ | Bayesian | Logistic regression |
| Algorithm | EM | MapReduce | – |
| Variants | SNPs and indels | SNPS and indels | SNPs and indels |
Table 2: variant callers comparison
Genome-wide association study (GWAS)
Genetic variants themselves do not tell any information about the potential association with individual phenotypes, which could be potentially associated with cancer risks [21]. Using the aforementioned pipeline, we can detect various genetic variants from NGS reads. The GWAS study asks if whether there are specific genetic variants found more often than expected in individuals with the phenotype of interest such as disease. The most common approach of GWAS studies is the case-control scenario, which compares two groups of individuals, for instance a normal group and a cancer group, to see whether the group is enriched with certain genetic variants.
There are two primary classes of phenotypes: categories (typically binary) or quantitative. From the statistical point of view, quantitative traits tend to be better as they improve the power to detect a genetic effect, and have a more explainable analysis results. For some diseases, the disease risk has been quantified. For example, High-density lipoprotein(HDL) and low-density lipoprotein(LDL) cholesterol levels are strong predictors of heart disease.
Therefore, the genetic variant association studies can be conducted by examining the relationship between genetic variations and these quantitative levels. Genetic variants that influence these levels are also easy to explain- for example, a unit change in LDL level per allele. Many other disease traits can’t be clearly quantified. In such circumstances, category labels are usually to represent ‘affected’ or ‘unaffected’. This makes the association studies much more difficult considering the enormous difference in measurement error associated with classifying individuals as either ‘case’ or ‘control’ versus precisely measuring a quantitative trait.
When a well-defined phenotype is chosen for a study population, the statistical analysis for the association between genetic variants and genotypes can begin. Quantitative traits are normally analyzed using generalized linear model methods such as the most widely used ANOVA. The null hypothesis of an ANOVA using a single SNP is that there is no difference between the trait mean of any genotype group. The p-value can be calculated based on such null hypothesis. The binary trait is generally analyzed using either contingency table. Contingency table tests examine and measure the deviation from independence that is expected under the null hypothesis that there is no association between the phenotype and genetic variants. The chi-square test and Fisher’s exact test are most widely used tests in this category. In addition to single-locus analyses, genome-wide association studies provide a great opportunity to examine interactions between genetic variants across the genome (Multi-locus analysis). Unlike the single-locus test, the multi-locus tests are much more complicated. A common strategy is to restrict investigation of SNP combinations to those fall with an established biological context such as pathway or protein family. Generally, a statistical method will be used to examine the significance of all potential SNP-SNP combinations in the GWAS dataset. A widely used multi-locus analysis tool in this type is INTERSNP [22]. There are also some widely used GWAS analysis toolkit such as plink [23], which integrated both single-Locus and multi-Locus analysis. Plink also integrated multi-correction and Stratified analysis, which makes it very comprehensive and powerful. The comparison between different GWAS analysis methods can be found in table.
| Test/Tool | ANOVA | Chi-Square | Fisher’s exact test | INTERSNP | plink |
| Availability | – | – | – | http://intersnp.meb.uni-bonn.de/ | http://zzz.bwh.harvard.edu/plink/ |
| Analysis Type | Single-Locus | Single-Locus | Single-Locus | Multi-Locus | Single-Locus and Mult-Locus |
| Trait | Quantitative | Binary | Binary | Binary and Quantitative | Binary and Quantitative |
Table 3: Genetic variant association tests/Tools
Conclusion
Next-generation sequencing is very powerful for identifying rare and de novo variants. The NGS reads mapping is crucial for variant calling followed. Therefore, choosing the appropriate read aligner and parameter is non-trivial for the genetic variation analysis. In this review, we compared a batch of commonly used read aligner to provide the readers some ideas of choosing read aligners and setting parameters. In this study, we have also compared a few widely used variant callers and GWAS study methods. With those discussions, readers can have an idea of the major components of the general pipeline of genetic variation studies: reads mapping, variant calling and genotype association. Besides, readers are also able to know the characteristics of different methods for each component and choose the appropriate methods and parameters based on their own specific application scenarios.
Conflict of interest
None
Acknowledgments
None
References
1. Siva N. 1000 Genomes project. Nature Publishing Group; 2008.1. Siva N. 1000 Genomes project. Nature Publishing Group; 2008.
2. Church GM. The personal genome project. Molecular systems biology. 2005;1(1).
3. Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome research. 1998;8(12):1229–1231.
4. Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nature genetics. 2008;40(8): 955–962.
5. Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nature reviews Genetics. 2011;12(11):745.
6. Do R, Kathiresan S, Abecasis GR. Exome sequencing and complex disease: practical aspects of rare variant association studies. Human molecular genetics. 2012;21(R1):R1–R9.
7. Pereira PCB, Melo FM, De Marco LAC, Oliveira EA, Miranda DM, Simoes AC, et al. Whole-exome sequencing as a diagnostic tool for distal renal tubular acidosis. Jornal de Pediatria (Versao em Portuguˆes). 2015;91(6):583–589.
8. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9(4):357–359.
9. Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14): 1754–1760.
10. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25(17):3389–3402.
11. Smith TF, Waterman MS. Identification of common molecular subsequences. Journal of molecular biology. 1981;147(1):195–197.
12. Yu X, Guda K, Willis J, Veigl M, Wang Z, Markowitz S, et al. How do alignment programs perform on sequencing data with varying qualities and from repetitive regions? BioData mining. 2012;5(1):6.
13. Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome research. 2008;18(11):1851–1858.
14. Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K, et al. SNP detection for massively parallel whole-genome resequencing. Genome research. 2009;19(6):1124–1132.
15. Martin ER, Kinnamon D, Schmidt MA, Powell E, Zuchner S, Morris R. SeqEM: an adaptive genotype-calling approach for next-generation sequencing studies. Bioinformatics. 2010;26(22):2803–2810.
16. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079.
17. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20(9):1297–1303.
18. Challis D, Yu J, Evani US, Jackson AR, Paithankar S, Coarfa C, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC bioinformatics. 2012;13(1):8.
19. Pirooznia M, Kramer M, Parla J, Goes FS, Potash JB, McCombie WR, et al. Validation and assessment of variant calling pipelines for next-generation sequencing. Human genomics. 2014;8(1):14.
20. Liu X, Han S, Wang Z, Gelernter J, Yang BZ. Variant callers for next-generation sequencing data: a comparison study. PloS one. 2013;8(9):e75619.
21. Jaeger E, Webb E, Howarth K, Carvajal-Carmona L, Rowan A, Broderick P, et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13. 3 influence colorectal cancer risk. Nature genetics. 2008;40(1):26–28.
22. Herold C, Steffens M, Brockschmidt FF, Baur MP, Becker T. INTERSNP: genome-wide interaction analysis guided by a priori information. Bioinformatics. 2009;25(24):3275–3281.
23. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81(3):559–57
Copyright
© This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/