J Med Discov (2017); 2(2):jmd17026; DOI:10.24262/jmd.2.2.17026; Received June 24th, 2017, Revised July 16th, 2017, Accepted July 16th, 2017, Published July 24th, 2017
Advances in the Application of Protein Mass Spectrometry to Skeletal Muscle Biology
Dapeng Chen1*, Rian Q. Landers-Ramos2, Davi A.G. Mázala3
1 Department of Chemistry and Biochemistry, University of Maryland, College park, MD, USA
2 Division of Gerontology and Geriatric Medicine, Department of Medicine, University of Maryland School of Medicine and the Baltimore Veterans Affairs Geriatric Research, Education and Clinical Center, Baltimore, MD, USA
3 Children’s National Health System, Children’s Research Institute, Center for Genetic Medicine Research, Washington DC, USA
* Correspondence: Dr. Dapeng Chen, Department of Chemistry and Biochemistry, University of Maryland, College Park, MD 20740 USA, cdp@umd.edu
Abstract
Protein mass spectrometry has been emerging as a powerful technology in the studies of human diseases. The advances in this technology, such as instrumentation, sample preparation, and bioinformatics data analysis, makes this technology more feasible to serve the scientific community. In this review, we will focus on the application of this technique in studies of skeletal muscle biology including skeletal muscle atrophy, muscular dystrophy, and skeletal muscle dysfunction-related metabolic syndromes. Particularly, we will introduce a system of methods used in protein mass spectrometry and discuss the scenarios in which protein mass spectrometry plays an essential role in protein characterization, such as posttranslational modifications and protein-protein interactions, and promoting our understanding of molecular mechanisms that cause skeletal muscle diseases. Finally, we will also discuss examples of successfully conducted protein mass spectrometry analysis to decipher complex molecular processes involved in skeletal muscle diseases.
Keywords: Protein, mass spectrometry, skeletal muscle biology.
Introduction
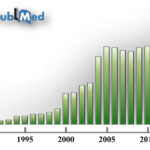
Mass spectrometry is one of the most accurate analytical techniques that provide the masses of molecules [1]. Due to its specificity and robust results, this technique has been emerging as an indispensable tool for large-scale protein characterization in biological research, clinical studies, and pharmaceutical manipulation [1]. This trend can be witnessed from the PubMed database of references and abstracts on the topics of life science and biomedical research. In 1990, only 11 papers were published on the topic of “protein mass spectrometry”; however, this number greatly increased to 450 in 2015 (Figure 1). Specifically, the number demonstrated a ~2-fold increase from 1999 to 2000 and from 2003 to 2004 presumably because of the improvement in protein database [2] and the advances in mass spectrometry instrumentation [3].
The human proteome includes about 100,000 proteins [4]. The complexity of the human proteome could be much more significant if protein posttranslational modifications (PTMs) such as phosphorylation, oxidation, glycosylation, methylation, and acetylation are considered. Alterations in protein patterns contribute to cellular disorders, tissue dysfunction, and human diseases [5]. Therefore, it is of great interest for scientists to observe these molecular alterations and interpret their biological consequences. Two protein mass spectrometry-based strategies are routinely used for global protein analysis: bottom-up proteomics and top-down proteomics [6, 7]. In bottom-up proteomics strategy, proteins are characterized by analysis of peptides that are produced from proteins with enzymatic digestion while top-down proteomics strategy directly analyzes intact proteins [6, 7]. Top-down proteomics strategy is unfavorable in global protein analysis due to fragmentation methods and its extremely poor capacity to handle larger proteins such as membrane proteins [8]. Therefore, this review will focus on the more-widely used bottom-up proteomics strategy for protein global analysis [6].
Skeletal muscle disorders are commonly found in a variety of pathological conditions such as skeletal muscle atrophy, muscular dystrophies, metabolic diseases, and cancers [9-20]. Astonishingly, there are limited available pharmaceutical inventions for the treatment of skeletal muscle diseases [14, 19]. For example, Riluzole (Rilutek) and Radicava (edaravone) are the only two U.S. Food and Drug Administration (FDA)-approved drug to treat patients with amyotrophic lateral sclerosis (ALS) [14], a fatal disease characterized by progressive muscle atrophy. Additionally, the FDA recently granted accelerated approval to Eteplirsen (an exon-skipping drug) and Emflaza or deflazacort (corticosteroid) for the treatment of Duchenne muscular dystrophy (DMD) [19]. DMD is characterized by progressive loss of muscle mass due to increased muscle damage and inflammation, and prednisone was the only FDA-approved treatment until recently [19].
The difficulty of discovering novel therapeutic targets for the treatment of various skeletal muscle diseases results from a poor understanding of the molecular mechanisms that contribute to disease pathogenesis. Protein mass spectrometry is extremely useful for this purpose and this notion has been discussed in several published scientific articles [21]. Therefore, in this review, we will discuss examples in which protein mass spectrometry has been used for analyzing changes in global protein in skeletal muscle biology. Specifically, we will discuss the usage of mass spectrometry for the evaluation of protein posttranslational modifications such as phosphorylation, acetylation, and glycosylation, protein-protein interactions, and protein quantitation. We will also briefly touch on some technical details in protein mass spectrometry analysis. It is our hope that this review can provide a useful guidance to scientists seeking to take advantage of this beautiful technique and promote their scientific ideas.
Figure 1: The numbers of articles published on the topic of “protein mass spectrometry” from 1990 to 2016. The data is acquired from PubMed on May 01, 2017. In 1990, only 15 papers were published in this topic. In 2015, this number increased to 437, representing a ~30-fold change.
Application of protein mass spectrometry analysis in studies of skeletal muscle diseases
Protein mass spectrometry technique
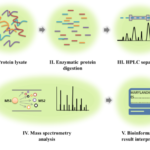
A typical platform in bottom-up proteomics method includes protein sample denaturing, enzymatic/chemical protein digestion, protein separation methods, mass spectrometry analysis, and bioinformatics (Figure 2) [6]. In the sample preparation step, proteins isolated from cells, tissues or cell supernatants are denatured, reduced, and alkylated. Essentially, during protein denaturing, proteins will be unfolded and the disulfide bonds will be reduced, providing access to proteases for the production of shorter peptides [6]. As the first step of the whole workflow protein denaturing is extremely important for the success of protein mass spectrometry analysis. Chemical detergents, such as sodium dodecyl sulfate (SDS) and tergitol-type NP-40, strongly interfere with mass spectrometry and should not be present in protein samples [6]. Poor protein sample preparation would result in protein clogging during protein separation with liquid chromatogram (LC) analysis, incomplete protein digestion, and low protein identification. After protein denaturing, protein digestion can be conducted with either proteases or chemicals. Although chemical-based protein digestion method has its own advantages, protease-assisted protein digestion is still the most widely used method for this purpose. Among these proteases, trypsin, which cleaves proteins preferably at the carboxyl side of the amino acid lysine (K) and arginine (R), has been the most efficient standard for protein digestion due to its robust, low cost, and high specificity. Other proteases, such as Asp-N and pepsin, are also available for the purpose of protein digestion. However, their usage for protein mass spectrometry analysis is hindered either due to their poor selectivity and the cost [6].
Reverse phase high-performance liquid chromatography (HPLC) is the principal method for peptide separation followed by mass spectrometry analysis [21]. The separation is completed by physical interactions between peptides and a solid adsorbent material (usually carbon 18 column), which causes different flow rates of peptides [21]. Protein separation by HPLC is the first step of sample simplification and very critical for mass spectrometry analysis. Recent years have witnessed the improvement in protein separation techniques such as capillary electrophoresis and ion mobility [22, 23]. Those methods are extremely useful for distinguishing different protein/peptide isoforms but their reproducibility and compatibility with mass spectrometry is still problematic [22, 23].
Figure 2: The workflow of bottom-up proteomics. This strategy includes five main steps: Protein sample preparation, protein digestion, HPLC separation, mass spectrometry analysis, and result interpretations with bioinformatics analysis. Protein samples are prepared from cells, tissues or cell lysates and treated with specific proteases. Peptides produced from the digestion step are then separated using HPLC and sent for the mass spectrometry analysis. Tandem mass spectrometry will be used to produce mass spectra, which can be interpreted with different bioinformatics tools for the purpose of peptide and protein identification.
Protein mass spectrometry analysis is completed by tandem mass spectrometry (MS/MS), which mainly includes two steps: mass spectrum production (MS1) and peptide fragmentation (MS2). During MS/MS, a peptide precursor, which is MS1, will be selected and proceed to fragmentation for the production of product ions (MS2). The mass spectrometry detector will then generate mass spectrum of MS2s, which will be used for the sequence determination of peptides. During mass spectrometry analysis, fragmentation method is the most important component. Currently, collision-induced dissociation (CID) is the most widely used method for ion fragmentation, in which peptides meet a cloud of neutral molecules in the gas phase and peptides break at their weakest bonds [24]. Higher-energy collisional dissociation (HCD), a CID technique specific to the analysis of proteins, is the most widely used method [24]. Other fragmentation methods, such as electron transfer dissociation (ETD) and ultraviolet photodissociation (UVPD) have also been invented and the combination of different methods will unquestionably prompt protein mass spectrometry analysis and protein identification [25, 26]. There are several tools that can be used for the bioinformatics analysis of MS2 and peptide sequencing. Particularly, during the bioinformatics analysis, MS2 spectra will be compared with theoretical mass spectra of predicted peptides that generated in silico from a defined protein database and peptides and proteins that reach statistical significance will be identified. By doing so, thousands of proteins could be identified by a single set of biological protein sample running. There are several well-established protein database searching engines, such as MASCOT SEQUEST, MaxQuant, and X!Tandem, which can be used to determine protein mutations, posttranslational modifications, and protein expression level [27, 28].
Protein quantitation with mass spectrometry analysis in skeletal muscle diseases
Changes in protein expression levels are associated with biological phenotypes. Therefore, it is of great interest to use a quantitative method for the investigation of proteome abundance and its biological consequences. Due to its high specificity, mass spectrometry-based protein quantitation is an ideal method for this purpose [1]. In this section, we will present two common strategies that are used for protein quantitation with mass spectrometry analysis: label-free spectral counting and isotopic label-based protein quantitation.
Label-free protein quantitation with mass spectrometry in skeletal muscle diseases
Label-free mass spectrometry quantitation is a method that measures relative protein expression levels [29]. In this strategy, protein quantitation is conducted either by precursor signal intensity or spectral counting numbers. This concept is based on the fact that the total number or intensity of peptides is relevant with the abundance of proteins and that the matched spectra to peptides can be compared statistically to reflect the protein abundance in different samples [29]. This method has been gaining more attention in the scientific community due to its low cost, easy sample preparation, and great capacity to couple with other workflows such as in-gel trypsin digestion. Here we will discuss several studies that successfully conducted label-free protein quantitation. More specifically, we will discuss the superiority of this method over traditional methods such as western blots and enzyme-linked immunosorbent assay (ELISA).
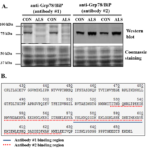
ALS is a neuromuscular disease characterized by progressive muscle wasting and atrophy [17]. It is generally supported that ALS is caused by the loss of nerves in the brain and spinal cord and that skeletal muscle atrophy is merely a consequence of motor neuron death. However, using a mouse model of ALS, we reported changes in skeletal muscle proteins prior to disease onset on ALS mice, suggesting that molecular factors within skeletal muscle could be the primary target of disease pathology [17]. Briefly, we evaluated if endoplasmic reticulum (ER) stress, which can be induced by protein aggregates accumulated in muscle cells, was activated in skeletal muscles of ALS mice [17]. For this purpose, we investigated the expression levels of several proteins involved in the ER stress pathway, including ER stress sensors PERK (protein kinase RNA-like endoplasmic reticulum kinase), IRE1 (inositol-requiring enzyme 1), protein disfulfide-isomerase (PDI), and glucose-regulated protein 78 kDa (Grp78/BiP). Our results demonstrated that Grp78/BiP showed different expression patterns when evaluated via immunoblotting (Figure 3A). Particularly, while one anti-Grp78/BiP antibody indicated a downregulation of the protein in ALS mice, a different anti-Grp78/BiP antibody showed an upregulation of the protein in ALS mice, which could be explained by the different binding sites of the two antibodies (Figure 3B). Considering the central role of Grp78/BiP in the activation of ER stress, we chose to use an unbiased and reliable method for a more precise analysis. In this case, we took advantage of protein mass spectrometry analysis and conducted an in-gel digestion coupling with label-free protein quantitation analysis. In this method, protein samples isolated from muscle tissues were separated by gel electrophoresis and protein bands at ~ 78 kDa were sliced and processed with in-gel trypsin digestion, which was then sent for the protein mass spectrometry analysis. The label-free method clearly showed that Grp78/BiP is upregulated in ALS mice compared to the control samples and this result is consistent with the observations of other ER stress markers such as PERK, IRE1, and PDI. This study undoubtedly showed that ER stress is activated in skeletal muscle of ALS and could contribute to cell death in muscle cells [17].
Another study that used the label-free strategy as an unbiased way to perform protein quantitation focused in evaluating serum irisin levels. [30]. Irisin is a product of fibronectin type III domain-containing protein 5 (FNDC5), which is coded by the FNDC5 gene [30]. A previous study suggested that irisin can be categorized as a myokine and presumably plays a very important role in exercise-promoted lipid metabolism [30]. Since the publication of the original study, arguments have been raised mostly challenging if this protein is really expressed in human serum since traditional analytical methods, such as immunoblotting and ELISA, could not provide a convincing evidence due to their non-specific protein binding issues [31]. To address the discussion in the literature, Jedrychowski and colleagues conducted a mass spectrometry-based detection of irisin in human serum [30]. Protein samples prepared from serum are notoriously complex due to the high abundance of albumin and immunoglobulin proteins and severe protein posttranslational modifications, which will cause an impediment in mass spectrometry analysis [30]. To deal with this issue, Jedrychowski and colleagues simplified protein serum samples with an affinity-based depletion and removed protein glycosylation using different de-glycosylated enzymes [30]. After protein sample simplification, the authors conducted an in-gel digestion on the protein bands where irisin was identified by immunoblotting and sent the digested peptides for mass spectrometry analysis [30]. For protein quantitation, Jedrychowski and colleagues chose several signature fragmentation ions produced from irisin-unique peptides. By comparing the intensity between their experimental samples and an isotopic internal standard peptide, the authors accurately determined the content of serum irisin and concluded that irisin increased significantly during exercising [30].
Figure 3: Western blot analysis of Grp78/BiP protein expression levels in skeletal muscle of ALS mice [16, 17]. A, Western blot analysis of Grp78/BiP protein in skeletal muscle of G93A* SOD1 ALS mice using two different antibodies. Protein loading was normalized with coomassie blue staining. The target Grp78/BiP protein bands showed around 75 kDa in control samples (CON) but not in ALS samples (ALS) using antibody #1. When using antibody #2, Grp78/BiP protein bands showed in both CON and ALS samples. B, Partial Grp78/BiP protein sequence (aa 410-644) and analysis of antibody binding sequence analysis of antibody #1 and #2 used in the study. The binding sequence of antibody #1 is from 570 to 600 (blue solid line) and the antibody #2 is from 525 to 628 (red dotted line).
The technical points in the study by Jedrychowski et al. include:
- Protein sample simplification strategy including removal of super-abundance non-relevant proteins, removal of posttranslational modifications for the purpose of protein mass accuracy, and protein separation with electrophoresis.
- Protein quantitation by taking advantage of characterized fragmentation ions generated from irisin-unique peptides and their reconstructed ion chromatogram.
III. The serum protein samples were unlabeled and an isotopic internal isotope-labeled peptide was used to conduct protein quantitation analysis for the absolute measurement of irisin.
- Protein quantitation analysis was conducted using selected reaction monitoring (SRM) method. In this method, a predefined mass to charge (m/z) value is isolated in the first quadruple mass spectrometer and undergoes collision-induced dissociation. Then, one specific fragmentation ion (product ion) is selected by the second mass spectrometer, which results in set of chromatograms that track this precursor ion to product ion transitions. The retention time and signal intensity for a specific transition are recorded and used for protein quantitation analysis.
Label-free protein quantitation with mass spectrometry analysis has been favored by the scientific community due to the low cost and simple sample preparation. However, in this strategy, different samples are analyzed 1) at different time points, 2) by different individuals, and 3) even using different analytical conditions, which give possibilities for instrumental and experimental bias. To address this issue, isotopic labeling with mass spectrometry analysis methods has been developed.
Isotopic labeling-based protein quantitation with mass spectrometry analysis
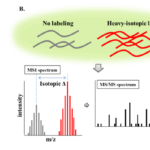
Isotopic labeling with mass spectrometry methods enables analysis of protein quantitation from different protein samples simultaneously, which overcomes the issue of experimental inconsistency [32]. Isobaric tags for relative and absolute quantification (iTRAQ) and stable isotope labeling with amino acid in cell culture (SILAC) are the two common isotope-based labeling methods for protein quantitation in mass spectrometry analysis [32-34]. In iTRAQ strategy, differential isotopically labeled tags are used as reporters and covalently attached to peptides during protein digestion (Figure 4A). Therefore, iTRAQ can be considered as an in vitro or on-bench sample preparation method. In iTRAQ analysis, several different samples (N≤8) can be prepared as a mixture and sent for the mass spectrometry analysis. During mass spectrometry analysis, the same peptides that are prepared from different samples have the same retention time will be analyzed, and peptide sequence information can be acquired from the patterns of fragmentation ions. The intensity of reporters can be used to indicate the relative abundance of the peptides.
iTRAQ strategy has been used for the investigation of protein carbonylation prepared from mitochondria from skeletal muscle [35]. Mitochondrial dysfunction has been reported to contribute to muscle pathology in several disease models such as skeletal muscle insulin resistance in type II diabetes. Meany and colleagues conducted a study in which iTRAQ proteomics strategy was used for the investigation of protein carbonylation in mitochondria isolated from rat skeletal muscle [35]. The authors developed the workflow that took advantage of protein enrichment and protein separation methods such as enrichment of mitochondrial proteins and affinity-based biotinylated protein isolation [35]. Furthermore, the authors evaluated several key technical aspects such as labeling efficiency and reproducibility. Results demonstrated 250 carbonylated proteins in addition to other posttranslational modifications such as phosphorylation and oxidation. This study by Meany et al. [35] provides the first survey about protein carbonylation in mitochondrial biology in skeletal muscle. Most importantly, the workflow presented in their study can be alternated to study other protein posttranslational modifications in a quantitative way [36].
One significant limitation of iTRAQ-based protein quantitation strategy is that isotope labeling is conducted after protein sample preparation and the labeling efficiency is the biggest concern [6]. Due to the competition during the labeling reaction, low abundance proteins could be suffering from very poor labeling and ignored during mass spectrometry analysis. In addition, an extra sample preparation step means further sample loss. Alternately, SILAC strategy has been developed for protein quantitation analysis for two purposes: 1) to improve labeling efficiency and 2) enable in vivo protein labeling (Figure 4B). In SILAC strategy, cells or animals are cultured either with a light isotopic labeled diet or a heavy-isotopic diet, which contains either carbon-13 (13C) or nitrogen-15 (15N). Depending on how many labeled amino acids a peptide contains, the mass difference can be calculated easily and the intensity between the light isotope-labeled and heavy isotope labeled peptides can be used to demonstrate the protein abundance between two groups (Figure 4). This method is also advantageous since the protein quantitation is determined in the precursor level and a complete fragmentation is not required. SILAC strategy has been used by Rayavarapu and colleagues to investigate the molecular mechanisms that contribute to skeletal muscle dysfunction in DMD, a genetic disorder caused by mutations in the dystrophin gene that affects all muscles [36]. In this study, the authors developed an SILAC study in which a mouse model of DMD was treated with heavy isotopic labeled diet and the proteome profiling was compared between control and disease models. The authors evaluated the efficiency of the labeling and the reproducibility of protein quantitation strategy (Figure 4C). In this way, the authors were able to conduct protein quantitation of a total of 897 proteins. Seventy-three proteins showed significant changes between wild-type vs. dystrophic mice and the bioinformatics analysis demonstrated that DMD is associated with changes in the integrin-linked kinase (ILK) signaling pathway as well as mitochondrial and metabolic enzymes [36].
Isotopic labeling-based protein quantitation has evolved as a very promising technique for proteome profiling. Other labeling methods have also been invented in addition to iTRAQ and SILAC, and such techniques play a very important role in answering important biological questions [1].
Figure 4: Schematic presentation of isotope labeling-based protein quantitation with mass spectrometry analysis. A, The strategy of protein quantitation using iTRAQ method. For example, four samples are labeled with four reporters (low masses). After mass spectrometry analysis, peptide sequence can be acquired from MS/MS analysis (grey bars). Protein quantitation information can be acquired from the intensity of four reporters (colored bars). B, The strategy of protein quantitation using SILAC method. The protein quantitation information is acquired from MS1 level. Since the peptides are labeling differently (light vs. heavy isotopes), the mass difference will be acquired in the MS1 spectra and the sequence information can be acquired from MS2 spectra. C, Mass spectra showing the precursor intensity of three proteins between disease and healthy animal models. The intensity of dystrophin between disease and health mouse models was compared and the mass spectrum showed that upregulation of dystrophin in health animals. GAPDH protein did not a show significant change and talin-1 was shown to be upregulated in disease animals.
Protein posttranslational modifications with mass spectrometry analysis in skeletal muscle diseases
The human proteome contains about 100,000 proteins. However, there are at least 300 types of post-translational modifications that have been identified physiologically and the complexity of the human proteome could be significantly greater if both numbers (proteome*PTMs) are combined (100,000*300!) [37-45]. Detection and identification of PTMs is extremely challenging and this leads to difficulties in the interpretation of their biological functions [38]. Mass spectrometry analysis enables large-scale protein characterization and has been emerging as the gold-standard for identifying PTMs. Detection of PTMs is based on the mass shift when a modification is added to a specific residue and mass spectrometry can detect this mass difference accurately (Figure 5A). For example, phosphorylation adds a mass of 80 Da while acetylation adds a mass of 42 Da. However, not all the mass shifts in PTMs are fixed and it demands extra experimental preparation when analyzing dynamic PTMs such as glycosylation (mass shift is between 200-2000 Da), which will be further discussed in this review [46]. PTMs are known to regulate various biological pathways and detection of their diversity would be essential to uncover disease pathological mechanisms in skeletal muscle diseases [14]. Below, we will discuss how protein mass spectrometry analysis methods have been used in skeletal muscle disease studies. Due to the large numbers of PTMs, we will only focus on protein phosphorylation, acetylation, and glycosylation.
Studies evaluating protein phosphorylation with mass spectrometry analysis
Mitochondrial dysfunction has been linked to cellular disorders and metabolic diseases including Type II diabetes [39]. It is of great interest to investigate alternated protein profiles in certain disease states and their functional consequences in mitochondria. Zhao and colleagues conducted a study in which the protein phosphorylation profiles in mitochondria were identified with mass spectrometry analysis. Since phosphorylated proteins are defined as low abundance proteins, an enrichment step, either targeting on a specific protein or phosphorylation, should be conducted in order to perform protein phosphorylation analysis [39]. In Zhao’s study, a titanium dioxide (TiO2) solid phase extraction method was used to enrich phosphorylated peptides prepared from skeletal muscle mitochondrial proteins [39]. By performing this phospho-proteomics study, the authors identified a total of 77 mitochondrial proteins with 155 phosphorylation sites on those proteins [39]. Cellular functional analysis showed that those proteins are involved in several key signaling pathways such as the tricarboxylic acid cycle and lipid metabolism. The study by Zhao et al [39] was the very first study in which phosphor-proteomics was conducted to reveal the molecular mechanisms that are associated with mitochondrial dysfunction in skeletal muscle insulin resistance [39]. More importantly, the workflow developed in the study has the potential to be applied in other experiments serving the same purpose of protein phosphorylation detection. Besides TiO2 column, other phosphorylation enrichment methods are available such as strong anion exchange and immobilized metal affinity chromatography [40].
Abnormal phosphorylation patterns have been reported to contribute to skeletal muscle dysfunction in insulin-resistant patients. Hojlund and colleagues conducted a large-scale study to characterize phosphorylated proteins in skeletal muscle of healthy human subjects [41]. In this study, protein samples were prepared from skeletal muscle and treated with trypsin. Digested peptides were then treated with strong cation exchange column and TiO2 for the enrichment of phosphorylation peptides. Mass spectrometry analysis on those enriched peptides identified 127 proteins containing 240 phosphoserines and 53 phosphothreonines. Functional analysis showed that those phosphorylated proteins play important roles in modulating sarcomeric function, excitation-contraction coupling, and glucose metabolism. Notably, they identified numerous novel phosphosites on sarcomeric Z-disk proteins, which would be very meaningful since these proteins are known to participate in skeletal muscle disorders [41].
Protein phosphorylation with mass spectrometry analysis can be also used for the study of phosphorylation patterns on specific proteins. For example, TBC1 Domain Family Member 4 (TBC1D4) protein is known to regulate glucose metabolism in skeletal muscle by regulating glucose transporter 4 (GLUT4) [42]. Typically, TBC1D4 phosphorylation is modulated by upstream kinases on serine and threonine residues [42]. However, it is speculated that TBC1D4 could be also phosphorylated by AMP-activated kinase (AMPK), a central protein kinase in regulating skeletal muscle metabolism. To investigate novel phosphorylation sites on this protein, Treebak and colleagues conducted a mass spectrometry analysis and observed several novel phosphorylation sites that could be regulated by AMPK [42]. By a critical evaluation of those sites, they found serine 711 (S711) to be of the greatest interest because of its agreement with AMPK phosphorylation motif. Finally, they confirmed the biological function of S711 as they showed that S711 phosphorylation is tightly linked to metabolic stimulus such as AMPK activator and exercising [42].
Studies evaluating protein acetylation with mass spectrometry analysis
Protein acetylation has received aggressive attention since the discovery of the protein family of sirtuins, which are NAD+-dependent deacetylases considered to have anti-aging effects [43]. Silent information regulator 1 (SIRT1) and 3 (SIRT3), which are regulated by the NAD+/NADH ratio, are expressed in skeletal muscle and regulate protein acetylation levels [43]. It has been reported that protein acetylation participates in various signaling pathways such as muscle cell differentiation, muscle metabolism, and stress response in skeletal muscle [44]. Lundby et al. performed a study evaluating protein acetylation patterns in different organs [45]. For the analysis, protein samples were prepared from different tissues such as brain, liver, cardiac muscle, and skeletal muscle. To facilitate protein acetylation detection with mass spectrometry analysis, acetylated peptides were enriched with anti-acetylation antibodies. A total of 4541 proteins containing 15474 acetylation sites were reported. Interestingly, it was found that more than 80% of proteins that belong to muscle contraction components were acetylated, with total of 941 acetylated proteins with 2811 acetylation sites identified in human skeletal muscle [45]. Further analysis showed that the highest abundance of protein acetylation is present in mitochondria and responsible for mitochondrial metabolic pathways. This could be highly meaningful since mitochondrial dysfunction has been linked to multiple skeletal muscle diseases [45].
Studies evaluating protein glycosylation with mass spectrometry analysis
Protein glycosylation is one of the most common PTMs and it has been estimated that 50% of mammalian proteins are glycosylated [46]. Protein glycosylation is known to participate in various biological events including cell-cell communication, intracellular trafficking, and cell membrane integrity [46]. However, the detection of protein glycosylation is extremely challenging mostly because unlike protein phosphorylation and acetylation showing a fixed mass change, the sugar structures are dynamic and the mass change of those structures can vary from 200 to 2000 Da [46]. Generally, two main types of glycoprotein are found in mammalian cells based on the link between carbohydrates and amino acids: O-linked and N-linked glycoproteins [46]. In the review, we will focus on the discussion on N-linked glycosylation with mass spectrometry analysis.
Peptide-N-Glycosidase F (PNGase F, Figure 5B), which is a native glycoaminidase enzyme cleaving the link between asparagine and N-acetylglycosamines, is used for the identification of N-linked glycosylation [47]. Generally, PNGase F initiates the transition from asparagine to aspartic acid and introduces a +0.98 Da mass shift, which can be analyzed with mass spectrometry for the determination of glycosites [47]. Recent studies showed the correlation between abnormal skeletal muscle protein glycosylation and metabolic disorders [14, 48]. For example, a number of studies showed that plasma cell antigen 1 (PC-1), which is a membrane glycoprotein, is either upregulated or hyperactive in skeletal muscle of insulin-resistant individuals and PC-1 overexpression both in vitro and in vivo contributes to impaired insulin receptor activity and its downstream signaling [48]. Besides PC-1, Glut-4, which plays a key role in glucose uptake in skeletal muscle, has been reported to undergo N-linked glycosylation [49]. Mutation-induced de-glycosylation was shown to disrupt Glut-4 intracellular trafficking and affect its quality control via impairing the protein turnover [49]. We conducted a study in which the patterns of N-linked glycosylation in skeletal muscle of Type 2 diabetic patients after aerobic exercise training (AEX) were investigated [14]. In this study, sedentary Type 2 diabetic patients completed a 12-month AEX program and skeletal muscle biopsy samples were obtained at baseline and at the completion of the traning program. Biopsies were treated with lectin-based glycoprotein enrichment and then analyzed by MS/MS. A total of 87 N-linked glycoproteins with 280 glycosites were identified in skeletal muscle of those biopsy samples [14]. By comparing spectral counts at baseline vs. after 12 months of AEX, we reported a reduction in the number of glycosylation sites in the isoform H17 of myeloperoxidase, which was associated with significant reduction in glycated hemoglobin (HbA1c) of 0.8 ± 0.1%. This study supports the notion that improved glycemic control is associated with alterations of skeletal muscle protein glycosylation patterns [14]. However, in this study, the physiological significance should be further evaluated and molecular mechanisms underlying how changes in protein glycosylation patterns affect glycemic control need further investigation.
Protein mass spectrometry analysis has been emerging as the most powerful tool to identify and quantify PTMs in biological samples. Recent years have witnessed the invention of enrichment methods for different PTMs, the advances in mass spectrometric instrumentation, and the improvement in the bioinformatics tools. Given these advances, identification of novel modified protein sites are expected to increase.
Protein-protein interactions with mass spectrometry analysis in skeletal muscle diseases
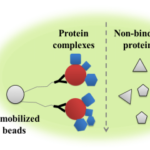
Protein-protein interactions (PPIs) is the basic component for the initiation of protein function and plays an essential role in regulating numerous key biological processes [50]. Abnormal PPIs results in cellular dysfunction and tissue pathology [50]. Therefore, understanding the biological function of PPIs is critical to revealing molecular factors involved in human diseases and the development of therapeutic targets. However, our current knowledge of PPIs in terms of characterizing interactions and elucidating their physiological consequences is still limited due to the low throughput and poor reproducibility of traditional techniques such as the yeast two-hybrid approach [50]. Mass spectrometry-based PPIs identification is advantageous due to its sensitivity, high-capacity, and accuracy. When coupled with protein immunoprecipitation technique, this technology can be used to characterize protein complexes in biological samples (Figure 6).
Mass spectrometry-based PPIs analysis has been used to investigate skeletal muscle pathology in ALS as our previous study showed that ER stress is activated in skeletal muscle of a mouse model of ALS [17]. In this study, Grp78/BiP, a key component in ER stress pathway, and its protein complexes were enriched with a co-immunoprecipitation method (co-IP) and analyzed with protein mass spectrometry [51]. Since the quality of the antibody is very important for co-IP, we developed a workflow to evaluate the reliability of the antibody used in the study. Results demonstrated that approximately 78 proteins interact with Grp78/BiP, including several calcium-regulating proteins and mitochondrial proteins [51]. We further conducted an in vitro assay to test the biological function of certain Grp78/BiP PPIs such as the interaction between Grp78/BiP and Sarco/endoplasmic reticulum Ca2+-ATPase 1 (SERCA1). SERCA1 is one of the most abundant proteins in skeletal muscle cells and is responsible for SR-calcium uptake during muscle relaxation [18]. Results demonstrated that the Grp78/BiP/SERCA1 interaction results in SERCA1 dysfunction which can lead to disruption in intracellular calcium homeostasis [51].
Interactome is defined as the identification of PPIs in a particular cell type and the interacting patterns of interactome can be mapped using mass spectrometry analysis [52]. The muscle regulatory factor (MRF) MyoD is an important transcriptional factor regulating skeletal myogenesis [53[. Boyarchuk and colleagues conducted a mass spectrometry analysis on the binding partners of MyoD to investigate factors involved in skeletal muscle terminal differentiation, which is critical to determine the epigenetic mechanisms contributing to the regulation of different muscle genes [53]. In the study, MyoD was tagged using a genetic method and its binding partners were enriched with a TAP-Tag purification strategy [53]. Several already known MyoD binding partners, such as pre-beta-cell leukemia transcription factor 1 (Pbx) and probable BOI—related E3 ubiquitin-protein ligase 2 (BRG2), were identified using this strategy. In addition, a number of novel important binding partners, such as (core-binding factor) CBF and beta cell-specific binding protein (EBB), were also identified [53].
Mass spectrometry-based PPIs analysis has been also used in the studies of dystrophin-glycoprotein complexes in DMD [54]. Murphy and colleagues conducted a study to characterize the dystrophin-glycoprotein complex in skeletal muscles of a mouse model of DMD using a pre-fractionation step and mass spectrometry analysis strategy [54]. A total of 281 proteins were identified, including dystrophin-associated proteins and factors associated with cellular metabolism, protein folding, and calcium regulation such as alpha-1-antitrypsin 1-5, palstin-2, galectin-3, and bone marrow proteoglycan [54].
Protein mass spectrometry-based analytical methods have been emerging as a very critical technique to identify PPIs. When coupled with bioinformatics tools, it can reveal factors that are associated with disease pathology and provide important messages on the development of therapeutic interventions and the discovery of biomarkers. It should be noted that a database of human protein complexes has been recently established (http://proteincomplexes.org/) [54]. This database contains > 4600 protein complexes, > 7700 proteins, and > 56000 unique interactions. The establishment of this powerful bioinformatics tool will significantly improve the understanding of the molecular mechanisms of human diseases and develop therapeutic targets for the treatment. Nevertheless, mass spectrometry-based PPIs analysis has been used extensively to resolve biological questions in other fields and the workflow can be adapted to study skeletal muscle biology.
Figure 5: Protein posttranslational modifications. A, Chemical representations of three PTMs: phosphorylation, acetylation, and glycosylation. The mass shifts of phosphorylation and acetylation are fixed but the glycosylation is dynamic, which could vary from 200 to 2000 Daltons. B, The mechanisms of PNGase F enzymatic deamidation. PNGase F initiates the transition of amino acid asparagine to aspartic acid, which provides a mass shift of 0.9840 Da, which can be detected with high-resolution mass spectrometry.
Figure 6: Enrichment of protein complexes with co-IP. The antibody is immobilized onto a solid base such as agarose or magnetic beads. The target protein and its binding complexes will bind to the antibody. After complete washing, non-binding proteins will be released and the target protein and the protein complexes can be collected after elution.
Conclusions and future directions
Protein mass spectrometry has been widely used by the scientific community to address critical biological questions. However, several limitations should be considered in this technique due to the high complexity of biological samples. Detection of abundance proteins by mass spectrometry is still very challenging. In real physiological conditions, proteins could have a very large dynamic range and, usually, the signals generated from high abundance proteins will dominate mass spectra, resulting in loss of the identification of low abundance proteins. This could be problematic for the detection of PTMs and an enrichment step is always required. The difficulties of detection of low abundance proteins require the improvement in every step of the workflow such as the improvement in protein separation techniques. For example, besides HPLC, other protein separation methods have been developed such as capillary electrophoresis and ion mobility [22, 23]. Those methods have been used in proteome studies and are of great advantages of detecting PTMs and protein isoforms. In addition, the advances in mass spectrometry, such as the improvement in mass spectrometer resolution, the fragmentation methods, and the combination of more than two mass spectrometers, has extensively promoted the numbers of proteins and the depth of protein identifications in biological samples. Nevertheless, protein mass spectrometry analysis has gained more attention in the scientific community and will be a very useful tool for the investigation of disease pathogenesis.
Acknowledgement
None
Conflict of Interest
None
References
1. Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198-207.
2. Desiere F, Deutsch EW, Nesvizhskii AI, Mallick P, King NL, Eng JK, et al. Genome Biol. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. 2005;6(1):R9.
3. Hardman M, Makarov AA. Interfacing the orbitrap mass analyzer to an electrospray ion source. Anal Chem. 2003; 75(7):1699-705.
4. Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509(7502):582-7.
5. Karve TM, Cheema AK. Small changes huge impact: the role of protein posttranslational modifications in cellular homeostasis and disease. J Amino Acids. 2011;2011:207691.
6. Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR 3rd. Protein analysis by shotgun/bottom-up proteomics. Chem Rev. 2013; 113(4):2343-94.
7. Michalski A, Damoc E, Lange O, Denisov E, Nolting D, Müller M, et al. Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol Cell Proteomics. 2012;11(3):O111.013698.
8. Reid GE1, McLuckey SA. ‘Top down’ protein characterization via tandem mass spectrometry. J Mass Spectrom. 2002;37(7): 663-75.
9. Mazala DA, English SA, Chen D, Chin ER. Alterations in intracellular free Ca2+ handling in mouse models for Duchenne muscular dystrophy. The FASEB Journal. 2011;25:lb599.
10. Jacobs B, Chen D, Chin ER. C2C12 cell culture model for investigating Ca2+-dependence of myocyte differentiation. The FASEB Journal. 2011;25(1 Supplement):lb598.
11. Chin ER, Goodman SR, Mazala DA, Chen D. Alterations in intracellular free Ca2+ concentrations in intact single muscle fibres from ALS mice. The FASEB Journal. 2011; 25(Supplement): 1051-49.
12. Chin ER, Mazala DA, Chen D. Alterations in Ca2+ regulatory proteins and Ca2+-dependent gene expression in skeletal muscle from ALS mice. The FASEB Journal. 2012;26(Supplement): 1075-6.
13. Mazala DA, Chen D, English SA, Grange RW, Chin ER. Effects of calpain inhibition in excitation-contraction coupling properties in dystrophic muscle exposed to fatiguing contractions. The FASEB Journal. 2012;26(Supplement):571-3.
14. Chen D, Wang Y, Prior SJ, Ryan AS, Ortmeyer HK, Blumenthal J, Beans J, Chin ER. Characterization of protein glycosylation in skeletal muscle of Type 2 diabetics after aerobic exercise training. The FASEB Journal. 2013;27(Supplement): lb703.
15. Chen D, Mazala D, Chin E. SERCA1 overexpression in skeletal muscle preserves motor function and delays disease onset in a mouse model of ALS. The FASEB Journal. 2015;29(1 Supplement):LB705.
16. Chen D, Mazala DA, English SA, Chin ER. BiP deficiency and ER stress in skeletal muscle of a mouse model of amyotrophic lateral sclerosis. The FASEB Journal. 2012;26(1 Supplement): lb783.
17. Chen D, Wang Y, Chin ER. Activation of the endoplasmic reticulum stress response in skeletal muscle of G93A* SOD1 amyotrophic lateral sclerosis mice. Front Cell Neurosci. 2015;9.
18. Chin ER, Chen D, Bobyk KD, Mazala DA. Perturbations in intracellular Ca2+ handling in skeletal muscle in the G93A* SOD1 mouse model of amyotrophic lateral sclerosis. Am J Physiol Cell Physiol. 2014;307(11):C1031-8.
19. Mázala DA, Pratt SJ, Chen D, Molkentin JD, Lovering RM, Chin ER. SERCA1 overexpression minimizes skeletal muscle damage in dystrophic mouse models. Am J Physiol Cell Physiol. 2015;308(9):C699-709.
20. Prior SJ, Goldberg AP, Ortmeyer HK, Chin ER, Chen D, Blumenthal JB, et al. Increased skeletal muscle capillarization independently enhances insulin sensitivity in older adults after exercise training and detraining. Diabetes. 2015;64(10):3386-95.
21, Mitulović G, Mechtler K. HPLC techniques for proteomics analysis—a short overview of latest developments. Brief Funct Genomics. 2006;5(4):249-60.
22. Simpson DC, Smith RD. Combining capillary electrophoresis with mass spectrometry for applications in proteomics. Electrophoresis. 2005;26(7‐8):1291-305.
23. McLean JA, Ruotolo BT, Gillig KJ, Russell DH. Ion mobility–mass spectrometry: a new paradigm for proteomics. Int J Mass Spectrom. 2005;240(3):301-15.
24. Sleno L, Volmer DA. Ion activation methods for tandem mass spectrometry. . Int J Mass Spectrom. 2004;39(10):1091-112.
25. Good DM, Wirtala M, McAlister GC, Coon JJ. Performance characteristics of electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2007;6(11):1942-51.
26. Ly T, Julian RR. Ultraviolet Photodissociation: Developments towards Applications for Mass‐Spectrometry‐Based Proteomics. Angew Chem Int Ed Engl. 2009;48(39):7130-7.
27. Tan Z, Nie S, McDermott SP, Wicha MS, Lubman DM. Single amino acid variant profiles of subpopulations in the MCF-7 breast cancer cell line. J Proteome Res. 2017; 20;16(2):842-51.
28. Tan Z, Yin H, Nie S, Lin Z, Zhu J, Ruffin MT, Anderson MA, Simeone DM, Lubman DM. Large-scale identification of core-fucosylated glycopeptide sites in pancreatic cancer serum using mass spectrometry. J Proteome Res. 2015;9;14(4):1968-78.
29. Zhu W, Smith JW, Huang CM. Mass spectrometry-based label-free quantitative proteomics. Biomed Res Int. 2009;2010.
30. Jedrychowski MP, Wrann CD, Paulo JA, Gerber KK, Szpyt J, Robinson MM, et al. Detection and quantitation of circulating human irisin by tandem mass spectrometry. Cell metabolism. 2015;22(4):734-40.
31. Timmons JA, Baar K, Davidsen PK, Atherton PJ. Is irisin a human exercise gene? Nature. 2012;488(7413):E9-10.
32. Han DK, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nature biotechnology. 2001;19(10):946-51.
33. Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7(3):340-50.
34. de Godoy LM, Olsen JV, de Souza GA, Li G, Mortensen P, Mann M. Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome biology.2006;7(6):R50.
35. Meany DL, Xie H, Thompson LV, Arriaga EA, Griffin TJ. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography‐stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7(7):1150-63.
36. Rayavarapu S, Coley W, Cakir E, Jahnke V, Takeda SI, Aoki Y, et al. Identification of disease specific pathways using in vivo SILAC proteomics in dystrophin deficient mdx mouse. Mol Cell Proteomics. 2013;12(5):1061-73.
37. Walsh CT, Garneau‐Tsodikova S, Gatto GJ. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl. 2005;44(45):7342-72.
38. Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nature methods. 2007;4(10):798-806.
39. Zhao X, León IR, Bak S, Mogensen M, Wrzesinski K, Højlund K, et al. Phosphoproteome analysis of functional mitochondria isolated from resting human muscle reveals extensive phosphorylation of inner membrane protein complexes and enzymes. Mol Cell Proteomics. 2011;10(1):M110-000299.
40. Mann M, Ong SE, Grønborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20(6):261-8.
41. Højlund K, Bowen BP, Hwang H, Flynn CR, Madireddy L, Geetha T, et al. In vivo phosphoproteome of human skeletal muscle revealed by phosphopeptide enrichment and HPLC− ESI− MS/MS. J Proteome Res. 2009;8(11):4954-65.
42. Treebak JT, Taylor EB, Witczak CA, An D, Toyoda T, Koh HJ, et al. Identification of a novel phosphorylation site on TBC1D4 regulated by AMP-activated protein kinase in skeletal muscle. Am J Physiol Cell Physiol. 2010;298(2):C377-85.
43. Imai SI, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24(8):464-71.
44. Mohamed JS, Wilson JC, Myers MJ, Sisson KJ, Alway SE. Dysregulation of SIRT-1 in aging mice increases skeletal muscle fatigue by a PARP-1-dependent mechanism. Aging (Albany NY). 2014;6(10):820.
45. Lundby A, Lage K, Weinert BT, Bekker-Jensen DB, Secher A, Skovgaard T, et al. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell reports. 2012;2(2):419-31.
46. Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nature biotechnology. 2003;21(6):660-6.
47. Dotz V, Haselberg R, Shubhakar A, Kozak RP, Falck D, Rombouts Y, et al. Mass spectrometry for glycosylation analysis of biopharmaceuticals. Trends Analyt Chem. 2015;73:1-9.
48. Youngren JF, Maddux BA, Sasson S, Sbraccia P, Tapscott EB, Swanson MS, et al. Skeletal muscle content of membrane glycoprotein PC-1 in obesity: relationship to muscle glucose transport. Diabetes.1996;45(10):1324-8.
49. Kandror K, Pilch PF. Identification and isolation of glycoproteins that translocate to the cell surface from GLUT4-enriched vesicles in an insulin-dependent fashion. J Biol Chem. 1994;269(1):138-42.
50. Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, et al. Large‐scale mapping of human protein–protein interactions by mass spectrometry. Mol Syst Biol. 2007;3(1):89.
51. Chen D, Wang Y, Chin ER. Identification of Grp78/BiP Protein Complexes Using Affinity Mass Spectrometry. 2017; American Society for Mass Spectrometry Conference.
52. Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180-3.
53. Boyarchuk E, Robin P, Fritsch L, Joliot V, Ait-Si-Ali S. Identification of MyoD Interactome Using Tandem Affinity Purification Coupled to Mass Spectrometry. J Vis Exp. 2016; 17(111):e53924.
54. Murphy S, Henry M, Meleady P, Zweyer M, Mundegar RR, Swandulla D, et al. Simultaneous pathoproteomic evaluation of the dystrophin-glycoprotein complex and secondary changes in the mdx-4cv mouse model of Duchenne muscular dystrophy. Biology. 2015;4(2):397-423.
Copyright
© This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/