Journal of Medical Discovery (2016); 1(1):16003; doi: 10.24262/jmd.1.1.16003; Received October 15th, Revised November 4th, Accepted November 8th, Published November 13th.
Genomic profile of sporadic colorectal cancer liver metastases versus primary tumors as defined by high-density microarray techniques
José María Sayagués1, Luís Antonio Corchete2, María Laura Gutiérrez1, Maria Eugenia Sarasquete2, María del Mar Abad3, Oscar Bengoechea3, Encarna Fermiñán4, María Fernanda Anduaga5, Sofia del Carmen3, Manuel Iglesias5, Carmen Esteban5, María Angoso5, Jose Antonio Alcazar5, Jacinto García5,*, Alberto Orfao1,*, Luís Muñoz-Bellvis5,*
1Cytometry Service-NUCLEUS, Department of Medicine, Cancer Research Center (IBMCC-CSIC/USAL) and IBSAL (University of Salamanca);
2Cáncer Research Center and Service of Hematology (University Hospital of Salamanca);
3Department of Pathology (University Hospital of Salamanca);
4Genomics Unit, Cancer Research Center (IBMCC-CSIC/USAL);
5Service of General and Gastrointestinal Surgery and IBSAL (University Hospital of Salamanca); Salamanca, Spain
*Jacinto García, Alberto Orfao and Luís Muñoz-Bellvis contributed equally to this work.
Prof. Alberto Orfao, Centro de Investigación del Cancer, Paseo de la Universidad de Coimbra S/N , 37007 Salamanca, Spain
Tel: +34-923294811, E-mail: orfao@usal.es
Currently, it is estimated that 90% of all sporadic colorectal cancer (sCRC) related deaths are (directly or indirectly) related to metastatic dissemination of the tumor (1). In fact, resection of localized lesions from primary non-metastatic tumors is associated with a favorable outcome, while in cases in whom metastases have occurred, complete cure is unlikely (2).
It is now well-established that the development and progression of any neoplasia is associated with an accumulation of tissue-specific and tumor-associated genomic alterations (3). In recent years, important advances have been achieved in sCRC tumors as regards the identification of their specific chromosomal abnormalities, as well as the precise patterns of intratumoral clonal evolution pathways associated with the metastatic process (4,5). Interestingly, primary tumors and their paired metastases frequently show many deregulated genes in common (6-9), suggesting that (e.g. liver) metastatic lesions originate from a tumor cell clone which is already present in the primary sCRC tumor and very closely related to it at the genomic level. However, the precise molecular changes that are associated with the metastatic process still remain to be completely identified. Therefore, identification of those specific genetic/genomic alterations that could be detected already at diagnosis and identify patients who are at risk of harboring (or developing) metastases, might significantly contribute to the development of new and useful strategies for the diagnosis and management of sCRC patients.
In the past, we have described those chromosomal abnormalities that are present in primary tumors from patients with metastatic vs. nonmetastatic sCRC, (10,11) using fluorescence in situ hybridization (FISH) and single nucleotide polymorphism (SNP) arrays (10,11). Briefly, FISH revealed several numerical (gains or losses of a whole chromosome) and structural chromosomal abnormalities, including del(17p) and del(22q), to be highly prevalent among patients with primary sCRC who had synchronous liver metastasis (11). In addition, high-resolution SNP-arrays provided further detailed information on those genetic alterations that are most frequently associated with metastatic sCRC. Among other alterations, these included numerical gains of chromosomes 8q, 13q, and 20q and losses of the 1p, 8p, 17p, 18q, and 22q chromosomal regions (10). More recently, several gene expression profiles (GEP) have been identified as predictors for stage II CRC patient outcome, using the Oncotype DX® Colon Cancer test (Genomic Health, Inc., Redwood City, CA) (12) and Coloprint® (Agendia, Inc., Irvine, CA) gene chips, among other gene expression profiling (GEP) platforms (13). However, once again, the molecular mechanisms underlying the association observed between such genomic profiles and metastatic colorectal cancer, remain largely unknown.
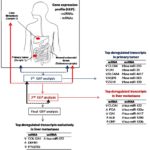
Sayagués et al investigated the molecular heterogeneity of sCRC based on simultaneous assessment of the overall GEP of both coding mRNA and non-coding RNA genes -including miRNA, small nucleolar and large intergenic RNAs-; in a group of 23 primary sCRC tumor samples and their paired liver metastases (14). To the best of our knowledge, so far, few studies have systematically analysed resected human metastatic tissues vs. their corresponding primary tumors, plus the associated normal colorectal tissue from the same patient (Figure 1) (7). Overall, the metastatic tumor samples analysed showed a GEP that was highly similar to that of their paired primary tumors. In fact, liver metastases systematically showed deregulated transcripts of those genes identified as being also altered in their paired primary colorectal carcinomas. Thus, both the primary tumors and their paired liver metastases showed overexpression of the FOXQ1, MMP7, CLDN1 and TACSTD2 mRNAS and the miR-4417, miR-503, miR-1290, miR-3687, miR-183, miR-224 and miR-1246 miRNAs, together with downregulated expression levels of the CLCA4, CA1, AQP8, ZG16, GUCA2B and SLC26A3 mRNAs and the amount of the miR-215, miR-133a, miR-375, miR-133b and miR-138 miRNAs.
Figure 1: Gene expression profiles (GEP) of primary vs. metastatic colorectal cancer. Scheme representing mRNA and miRNA differentially expressed between the different types of samples analyzed: primary sporadic colorectal tumors vs. normal colorectal tissue, colorectal liver metastases vs. normal colorectal tissue primary sporadic colorectal tumors vs. their paired liver metastases (q-values < 0.01).
Despite this, several coding and a fewer non-coding RNA transcripts were found to be specifically deregulated in liver metastases while expressed at normal levels in their paired primary tumors, which might potentially reflect adaption of the tumor cell to the liver microenvironment. Newly deregulated metastatic transcripts included overexpression of the APOA1, HRG, UGT2B4, RBP4 and ADH4 mRNAs and the miR-3180-3p, miR-3197, miR-3178, miR-4793 and miR-4440 miRNAs, together with decreased expression of the IGKV1-39, IGKC, IGKV1-27, FABP4 and MYLK mRNAS and the miR-363, miR-1, miR-143, miR-27b and miR-28-5p miRNAs. In addition, specific comparison between the GEP of liver metastasis and their paired primary tumor samples (paired analysis) revealed 52 mRNAS (14 down-regulated and 38 up-regulated genes) and two (over-expressed) miRNAs (miR-122 and miR-4322) to be differentially expressed between the two tumor tissues. If such results are confirmed in an independent series of metastatic sCRC the identified molecular markers for metastatic tumor might potentially be used to develop a multi-marker prognostic test (15). Interestingly most of the proteins (i.e.:MMP7, TACSTD2, CTHRC1 and KRT23) coded by this set of 52 differently expressed mRNA genes, have been found to be secreted and thereby present, both in tumor tissues and the plasma of CRC patients (16). These later observations not only demonstrated secretion of these proteins outside the tumor cell, but they also point out the potential utility of these genes as serum biomarkers for early diagnosis and monitoring of CRC patients.
In addition, Sayagués et al (9) also highlighted the activation of genes associated with the TGF-β signaling pathway in the metastatic tumors, -e.g. RHOA, SMAD2, SMAD4, SMAD5, SMAD6, BMPR1A, SMAD7 and MYC-; thereby, these genes also emerge as candidate genes to play an important role in CRC tumor metastasis. In fact, such findings have led to the speculation that TGFβ signaling might be responsible, at least in part, for the aggressiveness of metastatic sCRC tumors, by conferring a higher invasive phenotype, which is a prerequisite for the settlement and growth of distant metastasis (17).
In summary, Sayagués et al provide new insights about particular gene signatures potentially involved in sCRC metastasis also laying the path for the identification of novel biomarker candidates for both early diagnosis of sCRC tumors, and more efficient treatment and/or monitoring of CRC tumor metastasis.
Conflict of interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Acknowledgments
This work has been partially supported by grants from the Instituto de Salud Carlos III (ISCIII; Ministerio de Sanidad y Consumo, Madrid, Spain) (PI12/02053-FIS), Gerencia Regional de Salud de Castilla y León, Valladolid, Spain (GRS1302/A/16), Consejería de Sanidad (Junta de Castilla y Leon, Valladolid, Spain) (BIO/SA02/13), RTICC from the ISCIII (RD12/0020/0035-FEDER, RD12/0036/0048-FEDER), Fundación Memoria de Don Samuel Solórzano Barruso, (Salamanca, Spain) and Fundación Eugenio Rodríguez Pascual, (Madrid, Spain). JM Sayagués and ML Gutierrez are supported by grants (CES11/004 and PTA2014-09963-I) from the ISCIII, Ministerio de Economía y Competitividad, Madrid, Spain.
Reference
- Van CE, Cervantes A, Adam R, Sobrero A, van Krieken JH, Aderka D, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol 2016 Aug;27(8):1386-422.
- Weitz J, Koch M, Debus J, Hohler T, Galle PR, Buchler MW. Colorectal cancer. Lancet 2005 Jan 8;365(9454):153-65.
- Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet 2007 May;8(5):341-52.
- Sayagues JM, Fontanillo C, Abad MM, Gonzalez M, Sarasquete ME, Chillon MC, et al. Mapping of genetic abnormalities of primary tumours from metastatic CRC by high-resolution SNP arrays. PLoS ONE. 2010 Oct 29;5(10):e13752.
- Sayagues JM, Abad MM Barquero H, Gutierrez ML, Gónzalez-Gónzalez M, Jensen E, Bengoechea O, et al. Intratumoral cytogenetic heterogeneity of sporadic colorectal carcinomas suggests several pathways to liver metastasis. J Pathol. 2010 Jul;221(3):308-19.
- Mudduluru G, Abba M, Batliner J, Patil N, Scharp M, Lunavat TR, et al. A Systematic Approach to Defining the microRNA Landscape in Metastasis. Cancer Res 2015 Aug 1;75(15):3010-9.
- Lin AY, Chua MS, Choi YL, Yeh W, Kim YH, Azzi R, et al. Comparative profiling of primary colorectal carcinomas and liver metastases identifies LEF1 as a prognostic biomarker. PLoS ONE 2011;6(2):e16636.
- Vignot S, Lefebvre C, Frampton GM, Meurice G, Yelensky R, Palmer G, et al. Comparative analysis of primary tumour and matched metastases in colorectal cancer patients: evaluation of concordance between genomic and transcriptional profiles. Eur J Cancer 2015 May;51(7):791-9.
- Roessler S, Lin G, Forgues M, Budhu A, Hoover S, Simpson RM, et al. Integrative genomic and transcriptomic characterization of matched primary and metastatic liver and colorectal carcinoma. Int J Biol Sci 2015;11(1):88-98.
- Gonzalez-Gonzalez M, Fontanillo C, Abad MM, Gutierrez ML, Mota I, Bengoechea O, et al. Identification of a characteristic copy number alteration profile by high-resolution single nucleotide polymorphism arrays associated with metastatic sporadic colorectal cancer. Cancer 2014 Jul 1;120(13):1948-59.
- Gonzalez-Gonzalez M, Munoz-Bellvis L, Mackintosh C, Fontanillo C, Gutierrez ML, Abad MM, et al. Prognostic Impact of del(17p) and del(22q) as assessed by interphase FISH in sporadic colorectal carcinomas. PLoS ONE 2012;7(8):e42683.
- Kelley RK, Van Bebber SL, Phillips KA, Venook AP. Personalized medicine and oncology practice guidelines: a case study of contemporary biomarkers in colorectal cancer. J Natl Compr Canc Netw 2011 Jan;9(1):13-25.
- Kopetz S, Tabernero J, Rosenberg R, Jiang ZQ, Moreno V, Bachleitner-Hofmann T, et al. Genomic classifier ColoPrint predicts recurrence in stage II colorectal cancer patients more accurately than clinical factors. Oncologist 2015 Feb;20(2):127-33.
- Sayagues JM, Corchete LA, Gutierrez ML, Sarasquete ME, del Mar AM, Bengoechea O, et al. Genomic characterization of liver metastases from colorectal cancer patients. Oncotarget 2016 Sep 20.
- Maierthaler M, Benner A, Hoffmeister M, Surowy H, Jansen L, Knebel P, et al. Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. Int J Cancer 2016 Sep 15.
- Liu Y, Zhou J, Zhang C, Fu W, Xiao X, Ruan S, et al. HLJ1 is a novel biomarker for colorectal carcinoma progression and overall patient survival. Int J Clin Exp Pathol 2014;7(3):969-77.
- Peng X, Luo Z, Kang Q, Deng D, Wang Q, Peng H, et al. FOXQ1 mediates the crosstalk between TGF-beta and Wnt signaling pathways in the progression of colorectal cancer. Cancer Biol Ther 2015;16(7):1099-109.
Copyright
© This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/