Journal of Medical Discovery (2016); 1(1):JMD16005; Received October 18th , Revised November 5th , Accepted November 7th, Published November 14th.
Personalized oncology: the potential for tissue and cell-free DNA biopsies to capture tumor heterogeneity
Young Kwang Chae1,2,3, Andrew A. Davis2, Francis J. Giles1,2,3
1Developmental Therapeutics Program of Division of Hematology Oncology, Northwestern University Feinberg School of Medicine, Chicago, IL, USA,
2Department of Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, USA,
3Robert H. Lurie Comprehensive Cancer Center of Northwestern University, Chicago, IL, USA.
*Correspondence: Dr. Young Kwang Chae, Email: young.chae@northwestern.edu
A goal of personalized oncology is to adapt treatment in real-time in response to an individual patient’s dynamic genetic profile. Circulating tumor cell-free DNA (cfDNA) has emerged as an alternative technology to non-invasively monitor genomic mutations, copy number variants, and translocations in the peripheral blood. While multiple studies have reported high concordance when examining individual genes comparing tissue-based next generation sequencing with circulating tumor cfDNA, few prior studies had examined concordance of genomic alterations across multiple different genes.(1-4)
Our recently published work entitled “Concordance between genomic alterations assessed by next-generation sequencing (NGS) in tumor tissue or circulating cell-free DNA” compared mutational profiles of paired samples of tissue and tumor cfDNA in advanced solid tumors treated at our institution.(5) The goal of the work was to comprehensively examine concordance across a large number of genes, including all types of genomic alterations and variants of unknown significance. Our study reported a number of important findings. First, with existing technology, more mutations were detected in tissue as compared to cfDNA. Second, while concordance across all genes (including wild type/wild type genes) was high (>90%), concordance when genomic alterations were actually detected in either technique ranged from 11.8-17.1%. This has important clinical implications because one of the main goals of cfDNA assays is to detect specific resistance mutations. Third, unique mutations were discovered using both techniques with over 50% of mutations detected in one technique that were not detected using the other method. Potential reasons for our findings include tumor heterogeneity, inability to capture cfDNA at very low detection thresholds, fewer cfDNA variants released into the peripheral blood, our inclusion of a heterogeneous group of solid tumors, or different sequencing and detection techniques.
While our results were initially surprising, they highlight the complex biology of advanced solid tumors, which has served as both a challenge and an opportunity in personalized oncology. While the current gold standard for NGS is tissue, limitations exist with respect to taking a single biopsy from either a primary or metastatic site to capture the spatial heterogeneity and an evolving molecular profile of a tumor. There are also inevitable risks associated with repeat invasive biopsies.(6) In contrast, non-invasive emerging technologies including cfDNA, RNA sequencing, microRNA, circulating tumor cells (CTCs), and single cell proteomics and metabolomics have promising potential. These assays may enable monitoring genomic profile at various time intervals, non-invasively.
For cfDNA liquid biopsies, optimal timeframe regarding when and at what time intervals these assays should be performed remains unknown. In our retrospective study, median time interval between tissue and blood biopsies was approximately 90 days, but with a considerable range. While we hypothesized that concordance would be higher based on shorter time interval between tissue and blood biopsies, our study was unable to support this hypothesis. Previous work has reported that biopsies with at least one concordant mutation have a shorter timeframe between biopsies, but to our knowledge no studies have supported this consistently across many genes.(7) Future studies are needed to examine concordance at various intervals between biopsies and to monitor cfDNA over time.
While cfDNA assays theoretically have the potential to better represent tumor heterogeneity by capturing tumor DNA from multiple metastatic sites, the technique is predicated on the ability to detect circulating tumor DNA that is shed into the blood. The mechanism of tumor DNA being released into the blood is likely to occur via apoptosis and necrosis.(8) As a result, a hypothesis that needs to be tested is whether circulating tumor DNA preferentially represents therapy-sensitive tumor cells. While potentially advantageous for particular targeted therapies, a biological challenge would be to identify whether tumor DNA shed into the blood may not identify some resistant clones or subclones.(9) In many circumstances, therapy resistance is likely driven by these resistant clones, which may best be analyzed via other invasive techniques (repeat tissue biopsy, if feasible) or other non-invasive techniques (such as circulating tumor cells that are intact and have not undergone apoptosis).
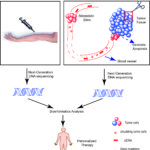
Recently, there have been two prominent editorial pieces in the New England Journal of Medicine and Nature pointing out how personalized oncology has failed to realize clinical benefits in many instances.(10,11) In the articles, the authors identified real challenges with regard to cost, tremendous individual tumor heterogeneity, and lack of data in randomized control trials that these approaches improve survival outcomes. Our study clearly supports the genetic tumor heterogeneity seen in tissue and blood. However, we envision the combination of tissue and non-invasive monitoring of peripheral blood cfDNA, RNA, CTCs, etc. will one-day enable more precise and dynamic monitoring for therapy to change in conjunction with tumor molecular evolution (Figure 1). In regards to the cost debate, an effort directed towards providing the right treatment to the right patient for the right duration of time based on the non-invasive assays discussed above may actually improve the value and cost-effectiveness of each therapy.
Figure 1. Model of DNA sequencing and comparison of cfDNA and tissue DNA.
Clearly, prospective trials examining whether utilizing circulating tumor cfDNA to guide treatment decisions can improve progression free or overall survival are needed. While our initial study compared concordance across a heterogeneous group of advanced solid tumors, we are currently examining concordance for particular histologies and at various time intervals between biopsy sampling and treatment, which may affect the degree with which cfDNA is shed into peripheral blood. Previous studies have demonstrated that different tumor types release DNA to different degrees in blood.(12) Therefore, we would expect concordance to vary as a result for certain tumor types. For now, there are emerging examples of circulating tumor cfDNA being capable of detecting resistance mutations, such as EGFR T790M in lung cancer or ESR1 in breast cancer.(13,14) To our knowledge, no prospective studies have identified whether detecting these alterations early in blood improves patient outcomes. However, certainly the potential exists.
Collectively, our study and many others highlight the substantial challenge and opportunity in personalized oncology. Given the considerable spatial and temporal heterogeneity of tumors, we must use a sophisticated combination of invasive and non-invasive techniques to monitor tumor evolution. We envision two exciting ways that non-invasive biopsies could change clinical practice. First, we suspect that the number of cfDNA clones and subclones, as well as somatic mutation frequency, may change prior to radiographic disease progression. In the future, this could enable changing therapy earlier based on an evolving genetic profile, rather than waiting for radiographic disease progression. Second, a long-term goal and incredible opportunity to improve survival would be for even earlier detection—detecting some marker of tumor cells, expression, or DNA in blood prior to metastatic disease. Certainly, there are tremendous challenges with respect to cost and when these non-invasive biopsies would be performed, but without further research into personalized oncology, these ambitious goals will never be achieved.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
None
References
- Mok T, Wu YL, Lee JS, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clinical cancer research : an official journal of the American Association for Cancer Research 2015;21:3196-203.
- Zill OA, Greene C, Sebisanovic D, et al. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. Cancer discovery 2015;5:1040-8.
- Kim ST, Lee WS, Lanman RB, et al. Prospective blinded study of somatic mutation detection in cell-free DNA utilizing a targeted 54-gene next generation sequencing panel in metastatic solid tumor patients. Oncotarget 2015;6:40360-9.
- Janku F, Angenendt P, Tsimberidou AM, et al. Actionable mutations in plasma cell-free DNA in patients with advanced cancers referred for experimental targeted therapies. Oncotarget 2015;6:12809-21.
- Chae YK, Davis AA, Carneiro BA, et al. Concordance between genomic alterations assessed by next-generation sequencing in tumor tissue or circulating cell-free DNA. Oncotarget 2016.
- Overman MJ, Modak J, Kopetz S, et al. Use of research biopsies in clinical trials: are risks and benefits adequately discussed? Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2013;31:17-22.
- Schwaederle M, Husain H, Fanta PT, et al. Use of Liquid Biopsies in Clinical Oncology: Pilot Experience in 168 Patients. Clinical cancer research : an official journal of the American Association for Cancer Research 2016.
- Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer research 2001;61:1659-65.
- Alix-Panabieres C, Pantel K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer discovery 2016;6:479-91.
- Tannock IF, Hickman JA. Limits to Personalized Cancer Medicine. The New England journal of medicine 2016;375:1289-94.
- Prasad V. Perspective: The precision-oncology illusion. Nature 2016;537:S63.
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Science translational medicine 2014;6:224ra24.
- Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2016;34:3375-82.
- Wang P, Bahreini A, Gyanchandani R, et al. Sensitive Detection of Mono- and Polyclonal ESR1 Mutations in Primary Tumors, Metastatic Lesions, and Cell-Free DNA of Breast Cancer Patients. Clinical cancer research : an official journal of the American Association for Cancer Research 2016;22:1130-7.
Copyright
© This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/