J Med Discov (2017); 2(4):jmd17039; DOI:10.24262/jmd.2.4.17039; Received September 12nd, 2017, Revised September 20th, 2017, Accepted September 23rd, 2017, Published September 28th, 2017
Broadly Neutralizing Antibodies (BNAbs): templates for HIV-1 vaccine design
Lixin Yan1,#, Lihong Liu2,#, Yilin Wang3,#, Xi Huang4,#, Qian Qian5, Jerica Lenberg5,6, Zheng Yang7, Shiqiang Lu8,*, Zehua Sun5,*, Rongxin Zhang9,*
1Harbin Medical University Affiliated 2nd Hospital, 246 Xuefu Road, Harbin, China, 150086.
2The Aaron Diamond AIDS Research Center. 455 First Avenue, New York, NY, 10016, USA. Email: lliu@adarc.org
3University of California, Irvine. 100 Pacific, Irvine, CA, 92618
4College of Foreign Studies, Hubei University of Automotive Technology, Email: viva124@connect.hku.hk
5Department of Medicine, National Jewish Health, 1400 Jackson Street, Denver, CO, 80206, USA.
6Augustana University, 2001 S Summit Avenue, Sioux Falls, SD, 571977
7Department of Tuberculosis Prevention, Shenzhen Center for Chronic Disease Control, Shenzhen, China
8Genscript USA Inc. 860 Centennial Ave. Piscataway, NJ 08854, USA. Email: shiqianglu@genscript.com
9Cancer Center Colorectal Department, Sun Yat-Sen University, Guangzhou, China, Guangzhou, China.
* Correspondence: Shiqiang Lu, PhD, Genscript USA Inc. 860 Centennial Ave. Piscataway, NJ 08854, USA. Email: shiqianglu@genscript.com; Zehua Sun, PhD, Department of Medicine, National Jewish Health. Denver, CO, United States, 80206. Email: zeh.sun@gmail.com; Rong-xin Zhang, PhD, Cancer Center Colorectal Department, Sun Yat-Sen University, Guangzhou, China. Email: zhangrx@sysucc.org.cn.
#The authors contribute equally to the work.
Abstract
For most licensed vaccines, protection efficacy is mainly conferred via the induction of neutralizing antibodies. Recently, potent and broadly neutralizing antibodies (bNAbs) have been isolated from certain HIV-1 elite controllers. The therapeutic and prophylactic efficacy of these bNAbs has been evaluated in animal models. The results were promising as bNAbs concentrations that could confer total protection were achievable by vaccination. Extensive efforts have been made to induce such bNAbs against HIV-1 infection, but none has succeeded yet. With a better understanding of the structure of bNAbs by X-ray diffraction and independent longitudinal observation of bNAbs in HIV-1 infected individuals, new ideas to guide the design of HIV-1 vaccines are expected to be proposed. Here, we reviewed strategies of viral escaping, characters and targets of HIV-1 bNAbs, and strategies of current vaccine design. Our review provides indication of potent bNAbs in role of vaccine design.
Keywords: broadly neutralizing antibodies (bNAbs), HIV-1, AIDS, vaccine design.
HIV-1 envelope and viral escape
HIV-1 envelope glycoprotein is not only responsible for viral binding with the facilitation of receptors and co-receptors, but also serves as the main target for HIV-1 specific antibody neutralization in controlling HIV-1 viremia and disease progress via antibody’s function in either impeding viral binding or blocking membrane fusion [1-9]. HIV-1 envelope is a transmembrane protein. The env gene of HIV-1 codes for the gp160 protein which forms a homotrimer. The gp160 precursor will be cleaved into gp120 and gp41 by the protease in the host cell. Thus the envelope protein is a trimer that is composed of three copies of heterodimers of gp41 and gp120. gp120 mediates viral binding with viral receptor CD4 and co-receptors CCR5/CXCR4, whereas gp41is responsible for membrane fusion [10]. HIV-1 specific antibodies induced in early stage of infection, which cannot control viral replication, are against gp41 [11,12]. After another 4 to 14 weeks, gp120-specific antibodies are induced, showing limited neutralizing ability against autologous viruses [13,14]. Meanwhile, through rapid viral replication and high error-prone reverse transcription, certain viral mutants resistant to autologous neutralizing antibodies quickly emerge and soon dominate the viral swarm [13,15]. The constant battle between neutralizing antibodies and escaped HIV-1 mutants finally leads to the appearance of bNAbs in certain long term non-progressors (LTNPs) at the cost of producing highly mutated HIV-1 viruses resistant to autologous bNAbs [14,16-20]. Recent progress in molecular structure has contributed greatly to the understanding of HIV-1 immune evasion which suggested an explanation that why HIV-1 envelope proteins fail to elicit effective neutralizing antibodies as other viral vaccines did [21,22]. The most common strategies recruited by HIV-1 in impeding bNAbs generation are concluded as follows:
Extensive glycosylation
Extensive glycosylation are commonly observed on envelopes of escaped viruses, which is speculated to be one of the main reasons for viral escape [13,23-29]. This “glycan shield” makes the epitopes on the envelope more inaccessible to antibodies. In addition, the glycan shield displayed on the envelope shares similarity to human glycoproteins, which has high risk to be not recognized by human B cells due to the immune tolerance to autologous antigens generated along with B cell development and maturation [30,31].
Conformational masking
Conformational masking is responsible for HIV-1 immune evasion [32]. Two functional sites on the envelope are the main beneficiaries to this strategy. One is CD4-induced binding site (CD4is), which appears on the envelope only after viral binding to CD4 receptor. The other is the membrane-proximal external region (MPER) region which is normally sealed off except when membrane fusion occurs [33]. The importance of conformational masking and heavy glycosylation in immune evasion has been well illustrated by the difference study in antibody immune responses during infection of HIV-1 and HIV-2. HIV-2 envelops are normally less conformational masked or glycosylated. Accordingly, bNAbs are commonly observed existing in HIV-2 infected individuals rather than HIV-1 infected individuals [34-38].
Paucity of HIV-1 spikes
The paucity of envelope molecules on the viral surface is another reason for envelope-induced immune evasion [39,40]. About 14 envelope spikes in average are expressed on each HIV-1viral particle, whereas the envelope spikes on other viruses are much higher [41]. Given that so few envelop spikes are displayed on the envelope protein, antibodies have difficulty in bivalent binding to two different spikes on the same viral particle. Thus, bivalent binding can hardly be achieved in HIV-1 which results the loss of efficacy.
HIV-1 epitopes targeted by bNAbs
Recently, an increasing number of bNAbs with improving breadth and potency have been identified from certain LTNPs [8,42]. In some cases, a single bNAb can recapitulate the neutralizing ability of serum [8,9,43-47]. But more often it resulted from the combined effects of multiple antibodies targeting different functional epitopes of HIV-1 envelopes [16,42,48-50]. Based on the antibody targeting activities, these functional epitopes can be classified: CD4 binding site, N-linked glycan-containing epitopes, and MPER.
CD4bs, a recessed pocket in HIV-1 envelope, is highly conserved which serves as a vulnerable site for interception by antibodies [51-54]. CD4bs antibodies can bind HIV-1 envelopes by mimicking CD4 via Complementarity-determining regions (CDR) H2 and adjacent residues [55-57]. Considering the special requirement for CDR H2, the variable regions of heavy chain (VH) of these CD4-mimic antibodies identified are invariably derived from either VH1-2 [44 ]or VH1-46 subfamily [49]. Most CD4-mimic antibodies identified from different LTNPs are from VH1-2 [44,48,49,55,58]. Recent studies showed that only VH1-2 germline encodes Trp50, Asn58, and Arg71, which are commonly existed in CD4-mimic antibodies, suggesting their critical role in antibody structure for envelope recognition. By contrast, limitations for light chains are few. Light chains with shorter CDR L1[56,58] or CDR L3 [59] are preferred. Compared with VH1-2 derived antibodies, VH1-46 antibodies, represented by NIH45-46, are less common. Besides, special requirements for CDR L3 and residues signature are unnecessary for VH1-46 antibodies [49,60]. In some cases, antibodies recognize CD4bs by using the long CDR H3 [61]. These antibodies exhibit less breadth and potency compared with CD4-mimic antibodies, which have been isolated from different LTNPs, including b12 [62] and CH103 [61].
N-linked glycan-containing epitopes
Although HIV-1 envelopes are generally covered with heavy glycans, certain bNAbs acquired the ability to recognize these potential N-glycosylation (PNG) sites after long-term evolution [43,63-66]. Given the high homogeneity of N-linked glycans on HIV-1 envelopes with host cellular proteins, development of glycan-specific antibodies should be especially difficult as the deletion of autoreactive B cells occurs early in vivo [67]. However certain bNAbs overcome this limitation by recognizing unique structures formed by multiple glycans or by both glycans and adjacent peptides that do not exist in cellular proteins. Asn332 glycan in V3 loop, along with conformational adjacent V3 domain, is the dominate epitope among all potential N-glycosylation (PNG) sites. Large numbers of HIV-1 glycan-specific antibodies are identified, such as PGT128 [43,68], PGT135 [43] and 2G12 [69]. Notably, the targeting epitope for PGT128 contains glycans at Asn301 and partial V3 loop beside Asn332 [68]. PGT128 can recognize and penetrate the glycan shield via its long CDR H2 and H3. The target epitope of 2G12 includes N-glycans at Asn295 and Asn339 in addition to Asn332. 2G12 can recognize the epitope via combining two antigen binding fragment (Fab) to a closely dimer [70]. Generally, Asn332-V3 antibodies are equally potent as CD4bs antibodies but with a narrower breadth [43,71]. The mechanism underlying its neutralizing activity remains unclear, but interference of CD4 binding may be involved [72].
As the first bNAb isolated by single B cell cloning in 2009, PG16 can recognize conformational epitopes which included glycans at Asn160 and adjacent residues [63,73]. More antibodies targeting similar epitope were subsequently isolated, such as PGT141-PGT145 [43] and CH01-CH04 [74]. The most remarkable character for these antibodies is the extremely long CDR H3. For example, CDR H3 of PG16 consists of 28 residues which can bind glycans with Asn156, Asn160, and part of the V2 loop [64]. Another study showed that the binding site of PG16 covers two gp120 on top of the envelope trimer [75], which explains why HIV-1 envelopes are not recognized by CD4 after PG16 binding [76]. However, the breadth of PG16 is not high and fails to neutralize some sensitive HIV-1 strains, possibly caused by the loss of L-linked glycans in the V3 loop [63]. Another HIV-1 glycan-specific antibody, 8ANC195 can recognize the N-linked glycans at Asn234 and Asn276, which is very close to CD4bs in terms of conformational structure [49].
MPER
Although gp41-specific antibodies dominate the early humoral immune response, bNAbs targeting MPER are few [77-81]. As an indispensable part of membrane fusion, MPER is relatively conserved. Many factors might contribute to the low frequency of MPER-specific antibodies isolated in vivo, such as, host mimicry [82-84], steric factor [85], and hydrophobicity [82]. In addition, MPER is partially buried and only transiently exposed [33,86]. MPER-specific antibodies are usually poly-reactive. MPER-specific B cells may be selectively deleted during B cell development and maturation process [65,67,87,88]. This concept was confirmed with the identification of human proteins that can cross-react with MPER-specific antibodies [84,89]. The deletion of MPER-specific B cells has also been observed during B cell development in 2F5 knock-in mice [90].
Characters of bNAbs against HIV-1
Extensive mutations
Nearly all bNAbs identified so far are characterized with extensive somatic mutations on both VH and VL fragments [8,91-96]. The average number of mutation nucleotides for mature human antibodies are between 15 and 20 [97], whereas HIV-1-specific bNAbs carry more mutations [42]. Notably, about 40-100 mutations are observed on VH for most bNAbs [43,44,49,55,63,71,98,99]. Later studies confirmed that these mutations are indispensable to the broad and potent neutralizing activity of bNAbs, because both binding and neutralization abilities of their corresponding germline antibodies are much weakened or even abolished [49,57,61,98,100]. Somatic mutations in vivo are induced by activation-induced cytidine deaminase (AID) in germinal center B cells [101]. Hot spot sequences are mainly located within CDR fragments that explained why more mutations are scattered in CDR regions than those in framework regions (FWR). Besides, single point mutations are more often observed, whereas insertion and deletion occurred less frequently [98,101]. Generally, FWRs have a certain degree of tolerance for mutations in DNA level and replacement of mutated FWR fragments with germline counterparts does not affect binding affinity [98]. Intriguingly, when the same replacement is examined in bNAbs, a significant reduction in neutralizing activity is observed, suggesting that the mutations in FWR fragments are critically important for neutralization [98].
Long CDR H3
CDR H3 is the most diverse sequence that is formed via VDJ recombination during the maturation of B cells [102-106] with an average length of 16 amino acids [97]. However, extremely short or long CDRs have been observed in a number of bNAbs [107]. For example, the length of CDR H3 is 30aa in PG9/PG16 [63,108-115] and 33aa in PGT145 [43]. Other bNAbs with extremely long CDR H3 include PGT121, PGT 135, 10E8, and CHO01-04 [107]. Notably, extremely long CDR H3 is not a prerequisite for all bNAbs, as its length in CD4-mimic antibodies is usually very short at around 5aa, which still shows broad and potent neutralizing activity [59].
Poly-reactivity
Poly-reactivity has been frequently observed in many bNAbs suggesting that these antibodies can recognize non-HIV-1 antigens. Given that most poly-reactive bearing B cells have been selectively deleted because of central and peripheral tolerance [67], it is interesting to explore why some HIV-1-specific antibodies can be rescued [65,82]. Some speculated that this phenomenon may be due to the paucity of envelops on HIV-1 particles which force bNAbs to seek another epitope for bivalent binding [40]. However, current evidence shows that poly-reactivity may be formed during the extremely long process of antibody maturation. Considering that most germline antibodies of bNAbs fail to bind with HIV-1 envelopes, B cells bearing these germline antibodies are possibly first activated by non-HIV-1 antigens [116,117].
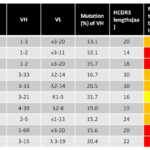
Figure 1 Notable characteristic of representative HIV-1 bNAbs
Design
Rational design of vaccines to elicit bNAbs
The importance of passive treatment with antibodies has been widely recognized. Compared with antiretroviral drugs, antibody therapy has extraordinary advantages [118-133]. First, antibody therapy provides long-term protection (two to four months) as the half-life of antibodies usually ranges from two to three weeks. Second, bNAbs with great breadth and potency are available, which can neutralize nearly 100% HIV-1strains in low concentration (0.05µg/ml). Third, given that HAART has failed to eliminate HIV-1 latency [134], the clearance of viral latent reservoirs by Fc fragment-induced immune response has been increasingly appreciated [135,136]. Antibody dependent cell mediate cytotoxicity (ADCC) helps eliminate intracellular HIV-1 virions by killing HIV-1 infected cells [137]. Fourth, certain bNAbs can inhibit cell-to-cell transmission of viral particles [138]. Finally, more effective viral control has been documented with the combined use of bNAbs and HAART according to the stages of viral replication cycle [139-142]. HIV-1 antibody therapies have been evaluated in animal models over a decade ago with the first generation HIV-1 neutralizing antibodies, b12, 2F5, and 2G12. The results were quite discouraging as protection was not observed [143]. Moreover, antibody-resistant HIV-1 mutants emerged quickly after treatment [143-145]. With the isolation of the secondary generation of bNAbs with extreme high neutralizing breadth and potency, the therapeutic efficacy of these bNAbs were evaluated in humanized mice and rhesus macaques [138,146-148]. Results showed that treatment with single bNAb can induce a transient drop in viral load, whereas treatment with combined antibodies induces long-term control of viral loads to undetectable levels [146]. Significantly, no antibody-resistant HIV-1 variants were observed during three months of treatment. Moreover, viremia rebounds did not occur after the withdrawal of antibodies in 10%-15% of humanized mice [146-148]. In addition, a recent study showed that bNAbs can effectively reduce the size of viral reservoirs in humanized mice [148]. Moreover, bNAbs were further examined in rhesus macaques. One shot of bNAbs provided rapid control of viral loads for nearly two months at about 5µg/ml [149,150]. Interestingly, studies showed that even single bNAbs can confer long-term control of HIV-1 load in most SIV infected rhesus macaques, compared with only transient reduction in humanized mice [149,150]. The reason why bNAbs provide better control in rhesus macaques than in humanized mice remains unclear. However, the difference in immune systems may be the main reason [140]. Therefore, although the efficacy of bNAbs therapy in human needs to be further verified, the preliminary data in animal models are promising and provide hope for fighting against HIV-1/AIDS in the future.
Potent bNAbs show great prophylactic or therapeutic applications in animal models [149-152]. Vaccines that can elicit such bNAbs would be greatly valuable for the finally eradication of HIV-1/AIDS. However, previous vaccine trials tested to date failed to induce such bNAbs in both animal models and humans [153]. An increasing number of bNAbs have been identified and characterized from HIV-1-infected individuals, so how bNAbs are naturally generated within the patients deserve further investigation, which may provide valuable clues for HIV-1 vaccine design. Both host and virus factors are speculated to be responsible for the generation of bNAbs [154]. If host factors are the main reason, designing vaccines to induce bNAbs in the majority of the immunized population may be impossible. However, recent studies proved that viral factors, especially the successive emergence of HIV-1 envelopes from escaped variants, may contribute to the elicitation and maturation of bNAbs. Previous studies from rhesus macaques indicated that the envelopes of founder viruses are associated with the initiation of bNAbs because SIVs with different envelopes show distinct abilities to induce neutralizing antibodies. For example, antibodies are induced in most macaques infected with AD8 but not DH12 virus [155,156]. The importance of the initial envelope was further approved by the natural occurrence of bNAbs in an individual with HIV-1 superinfections [157]. The individual was infected with HIV-1 viruses of two different lineages. However the neutralizing antibodies were induced by the viruses in one lineage. The diversity of a viral population may be associated with the breadth of neutralizing antibodies [158,159]. Based on these findings, two vaccine strategies have been proposed: 1) HIV-1 immunogen design based on the structure of bNAbs and targeting epitopes [160,161]; 2) reproducing the natural occurrence of bNAbs in vivo by multiple successive immunizations with correlated immunogens [42,50,61,162].
Design of immunogens based on neutralizing epitopes of envelops
The HIV-1 envelope susceptible sites to bNAbs have been studied for long. Thus, modified HIV-1 envelopes and their structure mimicals have been designed and tested in animal models. Unexpectedly, bNAbs failed to be induced by this strategy [160,163]. A later study reported that the inferred germline antibodies of most known bNAbs do not bind envelopes at all, which explains why the previous HIV-1 immunogens failed to induce bNAbs [49,57,98,100]. The germline precursors of 2F5 can bind envelopes, but they cross react with autologous proteins [164]. Thus, B cells expressing 2F5 germline antibody are most likely to be removed in the process of B cell development [67,82-84].
Design of immunogens based on the inferred germline precursors of bNAbs
The recombinant envelopes and peptides that can bind the germline antibodies by mimicking CD4bs and V1/V2 domains, respectively, have been designed for vaccination [165-167]. Theoretically, vaccination with these peptides may activate the expansion of CD4bs- or V1/V2- specific B cells. However, these trials failed because activation of B cells was insufficient. In addition, the frequency of mutations in variable regions of antibodies during B cells maturation in germinal center is high, which might be important for the neutralizing activity of bNAbs [101].
Design of immunogens based on B cell lineage maturation pathway
When mutations in mature antibodies induced by viral or bacterial infection reach to a certain level, they cannot be induced by additional stimulations [97,168-170]. Intriguingly, about 40-100 mutations accumulate in bNAbs. To explain this phenomenon, a coevolution model of viruses and antibodies has been proposed [42,49,50,61,171]. According to theory, early antibodies are elicited by the founder HIV-1 strains, which usually weakly bind envelopes of the founder viruses with low or even no neutralizing activity. After several rounds of antibody maturation in germinal center, these intermediate antibodies acquire better binding and neutralizing abilities against the early viral strains. Meanwhile, the diversity of the HIV-1 viral pool expands quickly because of the error-prone reverse transcriptase and relatively short life cycle of HIV-1. HIV-1 variants resistant to contemporaneous antibodies are selected in vivo because slight alterations in binding epitopes on envelope might exempt the virus to be neutralized by strain-specific antibodies produced in the early stages. Responding to the evolution of viruses, strain-specific antibodies also experience further mutations during affinity maturation in germinal center. Mutations at certain positions may help the antibodies to acquire additional breadth, which can neutralize both the founder and escape variants. Finally, bNAbs might appear in some HIV-1-infected individuals after the iterative repetitions of antibody mutations and HIV-1 escape. The long process of coevolution explains why bNAbs emerged after two to four years [98,171]. The elicitation and maturation of bNAbs were observed in certain HIV-1-infected individuals by two independent studies. The formation of CD4bs-specific bNAbs CH103 was first reported by Liao’s group in 2013 [61], whereas a similar antibody maturation pathway was observed in V1/V2-specific bNAbs by Doria-Rose and colleagues one year later [157]. As exemplified in the maturation pathway of the CH103 family, the germline precursor of unmutated ancestor antibodies shows no detectable binding activity with HIV-1 envelopes. However, early intermediate antibodies with limited mutations that can bind HIV-1 envelopes emerged about 14 weeks after infection, which has low binding affinity at 96,500nM. Since then the coevolution of HIV-1 envelopes and antibodies is longitudinally observed. The maturation affinity of CH antibodies was noted to increase from 96,500 nM for early intermediate antibodies to 2.4 nM for mature ones. Consistent with these observations, a new strategy was proposed to elicit bNAbs in individuals by mimicking the natural occurrence and maturation of human B cells, named B cell lineage immunogen design [162]. In brief, this strategy can be divided into three steps: 1) to isolate bNAbs and corresponding intermediate precursors from LTNPs. 2) to draw the phenogenic relationship of the unmutated ancestor antibody (UA), intermediate antibodies and mature antibodies. 3) To use these antibodies as templates for designing immunogens with high binding affinity. Unlike classical immunization schemes, distinct but closely related immunogens are used for prime and boost in B cell lineage vaccination. To induce the further maturation of antibodies, multiple rounds of continuing boosts are required [74,107,172-176].
Conclusion
In summary, the most encouraging progress came from the discovery of bNAbs from HIV-1 LTNPs with single B cell cloning technique in the past few years. These antibodies can efficiently reduce viral loads and slow down the disease progress as shown in humanized mice and rhesus macaques at the concentration accessible by vaccination, thereby providing hope for the design of HIV-1 immunogens. Different strategies for immunogen designs have been proposed. However HIV-1 vaccines that can elicit such bNAbs in vivo are still presently unavailable. Our review provides indications in potent neutralizing antibodies development and vaccine design against HIV-1.
Conflict of interest
The authors contribute equally to the manuscript and claimed no interest.
Acknowledgments
We wish to thank Dr. Mei-yun Zhang, Dr. Zhiwei Chen (The University of Hong Kong), and Dr. Paul Zhou (Institute Pasteur of Shanghai CAS) for helpful discussion. This research is no grant supported.
References
- Lewis, G. K., Pazgier, M. & DeVico, A. L. Survivors Remorse: antibody-mediated protection against HIV-1. Immunol Rev 275, 271-284, doi:10.1111/imr.12510 (2017).
- Lewis, G. K. The first 24 h: targeting the window of opportunity for antibody-mediated protection against HIV-1 transmission. Curr Opin HIV AIDS 11, 561-568, doi:10.1097/COH.0000000000000319 (2016).
- Schoofs, T. et al. HIV-1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV-1. Science 352, 997-1001, doi:10.1126/science.aaf0972 (2016).
- Hessell, A. J. et al. Achieving Potent Autologous Neutralizing Antibody Responses against Tier 2 HIV-1 Viruses by Strategic Selection of Envelope Immunogens. J Immunol 196, 3064-3078, doi:10.4049/jimmunol.1500527 (2016).
- Verkoczy, L. Humanized Immunoglobulin Mice: Models for HIV Vaccine Testing and Studying the Broadly Neutralizing Antibody Problem. Adv Immunol 134, 235-352, doi:10.1016/bs.ai.2017.01.004 (2017).
- Kelsoe, G. & Haynes, B. F. Host controls of HIV broadly neutralizing antibody development. Immunol Rev 275, 79-88, doi:10.1111/imr.12508 (2017).
- Bonsignori, M. et al. Antibody-virus co-evolution in HIV infection: paths for HIV vaccine development. Immunol Rev 275, 145-160, doi:10.1111/imr.12509 (2017).
- Sun, Z., Li, J., Hu, X., Shao, Y. & Zhang, M. Y. Reconstitution and characterization of antibody repertoires of HIV-1-infected “elite neutralizers”. Antiviral Res 118, 1-9, doi:10.1016/j.antiviral.2015.02.006 (2015).
- Sun, Z., Lu, S., Yang, Z., Li, J. & Zhang, M. Isolation and characterization of HIV-1 envelope glycoprotein specific B cell from immortalized human naive B cell library. J Gen Virol, doi:10.1099/jgv.0.000706 (2017).
- Knipe, D. M. & Howley, P. M. Fields Virology 6th Edition. (2013).
- McMichael, A. J., Borrow, P., Tomaras, G. D., Goonetilleke, N. & Haynes, B. F. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol 10, 11-23, doi:10.1038/nri2674 (2010).
- Tomaras, G. D. et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol 82, 12449-12463, doi:10.1128/JVI.01708-08 (2008).
- Wei, X. et al. Antibody neutralization and escape by HIV-1. Nature 422, 307-312, doi:10.1038/nature01470 (2003).
- Mikell, I. et al. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog 7, e1001251, doi:10.1371/journal.ppat.1001251 (2011).
- Richman, D. D. et al. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol 68, 1660-1666 (1994).
- Moore, P. L. et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med 18, 1688-1692, doi:10.1038/nm.2985 (2012).
- Doria-Rose, N. A. et al. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J Virol 84, 1631-1636, doi:10.1128/JVI.01482-09 (2010).
- Dimonte, S. Different HIV-1 env frames: gp120 and ASP (antisense protein) biosynthesis, and theirs co-variation tropic amino acid signatures in X4- and R5-viruses. J Med Virol 89, 112-122, doi:10.1002/jmv.24611 (2017).
- Pouran Yousef, K. et al. Inferring HIV-1 Transmission Dynamics in Germany From Recently Transmitted Viruses. J Acquir Immune Defic Syndr 73, 356-363, doi:10.1097/QAI.0000000000001122 (2016).
- Pham, H. T., Mesplede, T. & Wainberg, M. A. Effect on HIV-1 viral replication capacity of DTG-resistance mutations in NRTI/NNRTI resistant viruses. Retrovirology 13, 31, doi:10.1186/s12977-016-0265-x (2016).
- Paquin-Proulx, D. et al. Innate Invariant NKT Cell Recognition of HIV-1-Infected Dendritic Cells Is an Early Detection Mechanism Targeted by Viral Immune Evasion. J Immunol 197, 1843-1851, doi:10.4049/jimmunol.1600556 (2016).
- Rolland, M. HIV-1 immune evasion-a threat to effective vaccines? Nat Med 22, 580-581, doi:10.1038/nm.4119 (2016).
- Sagar, M., Wu, X., Lee, S. & Overbaugh, J. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J Virol 80, 9586-9598, doi:10.1128/JVI.00141-06 (2006).
- van Gils, M. J. et al. Longer V1V2 region with increased number of potential N-linked glycosylation sites in the HIV-1 envelope glycoprotein protects against HIV-specific neutralizing antibodies. J Virol 85, 6986-6995, doi:10.1128/JVI.00268-11 (2011).
- Nelson, C. S. et al. Combined HIV-1 Envelope Systemic and Mucosal Immunization of Lactating Rhesus Monkeys Induces a Robust Immunoglobulin A Isotype B Cell Response in Breast Milk. J Virol 90, 4951-4965, doi:10.1128/JVI.00335-16 (2016).
- Musich, T. & Robert-Guroff, M. New developments in an old strategy: heterologous vector primes and envelope protein boosts in HIV vaccine design. Expert Rev Vaccines 15, 1015-1027, doi:10.1586/14760584.2016.1158108 (2016).
- de Taeye, S. W., Moore, J. P. & Sanders, R. W. HIV-1 Envelope Trimer Design and Immunization Strategies To Induce Broadly Neutralizing Antibodies. Trends Immunol 37, 221-232, doi:10.1016/j.it.2016.01.007 (2016).
- Muhle, M., Kroniger, T., Hoffmann, K. & Denner, J. The immunosuppressive domain of the transmembrane envelope protein gp41 of HIV-1 binds to human monocytes and B cells. Immunol Res 64, 721-729, doi:10.1007/s12026-015-8776-4 (2016).
- Sliepen, K. & Sanders, R. W. HIV-1 envelope glycoprotein immunogens to induce broadly neutralizing antibodies. Expert Rev Vaccines 15, 349-365, doi:10.1586/14760584.2016.1129905 (2016).
- Rawat, P. & Spector, S. A. Development and characterization of a human microglia cell model of HIV-1 infection. J Neurovirol 23, 33-46, doi:10.1007/s13365-016-0472-1 (2017).
- Trejbalova, K. et al. Development of 5′ LTR DNA methylation of latent HIV-1 provirus in cell line models and in long-term-infected individuals. Clin Epigenetics 8, 19, doi:10.1186/s13148-016-0185-6 (2016).
- Kwong, P. D. et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678-682, doi:10.1038/nature01188 (2002).
- Frey, G. et al. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci U S A 105, 3739-3744, doi:10.1073/pnas.0800255105 (2008).
- de Silva, T. I. et al. Potent autologous and heterologous neutralizing antibody responses occur in HIV-2 infection across a broad range of infection outcomes. J Virol 86, 930-946, doi:10.1128/JVI.06126-11 (2012).
- Ozkaya Sahin, G. et al. Potent intratype neutralizing activity distinguishes human immunodeficiency virus type 2 (HIV-2) from HIV-1. J Virol 86, 961-971, doi:10.1128/JVI.06315-11 (2012).
- Devadas, K. et al. Identification of Host Micro RNAs That Differentiate HIV-1 and HIV-2 Infection Using Genome Expression Profiling Techniques. Viruses 8, doi:10.3390/v8050121 (2016).
- Steiniche, D. et al. Diabetes mellitus and impaired fasting glucose in ART-naive patients with HIV-1, HIV-2 and HIV-1/2 dual infection in Guinea-Bissau: a cross-sectional study. Trans R Soc Trop Med Hyg 110, 219-227, doi:10.1093/trstmh/trw017 (2016).
- Hollenbaugh, J. A., Montero, C., Schinazi, R. F., Munger, J. & Kim, B. Metabolic profiling during HIV-1 and HIV-2 infection of primary human monocyte-derived macrophages. Virology 491, 106-114, doi:10.1016/j.virol.2016.01.023 (2016).
- Klein, J. S. & Bjorkman, P. J. Few and far between: how HIV may be evading antibody avidity. PLoS Pathog 6, e1000908, doi:10.1371/journal.ppat.1000908 (2010).
- Mouquet, H. et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature 467, 591-595, doi:10.1038/nature09385 (2010).
- Zhu, P. et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441, 847-852, doi:10.1038/nature04817 (2006).
- Scheid, J. F. et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636-640, doi:10.1038/nature07930 (2009).
- Walker, L. M. et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466-470, doi:10.1038/nature10373 (2011).
- Wu, X. et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856-861, doi:10.1126/science.1187659 (2010).
- Szymanek-Pasternak, A., Szymczak, A., Zalewska, M., Malyszczak, K. & Knysz, B. Risk factors for chronic kidney disease do not influence the serum levels of asymmetric dimethylarginine in HIV-1-infected patients without significant renal disease. Pol Arch Med Wewn 126, 672-680, doi:10.20452/pamw.3508 (2016).
- Boadu, R., Darko, G., Nortey, P., Akweongo, P. & Sarfo, B. Assessing the sensitivity and specificity of First Response HIV-1-2 test kit with whole blood and serum samples: a cross-sectional study. AIDS Res Ther 13, 9, doi:10.1186/s12981-016-0092-0 (2016).
- Khattar, S. K. et al. Mucosal Immunization with Newcastle Disease Virus Vector Coexpressing HIV-1 Env and Gag Proteins Elicits Potent Serum, Mucosal, and Cellular Immune Responses That Protect against Vaccinia Virus Env and Gag Challenges. MBio 6, e01005, doi:10.1128/mBio.01005-15 (2015).
- Georgiev, I. S. et al. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 340, 751-756, doi:10.1126/science.1233989 (2013).
- Scheid, J. F. et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633-1637, doi:10.1126/science.1207227 (2011).
- Wibmer, C. K. et al. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS Pathog 9, e1003738, doi:10.1371/journal.ppat.1003738 (2013).
- Kwong, P. D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648-659, doi:10.1038/31405 (1998).
- Huang, J. et al. Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth. Immunity 45, 1108-1121, doi:10.1016/j.immuni.2016.10.027 (2016).
- Wibmer, C. K. et al. Structure of an N276-Dependent HIV-1 Neutralizing Antibody Targeting a Rare V5 Glycan Hole Adjacent to the CD4 Binding Site. J Virol 90, 10220-10235, doi:10.1128/JVI.01357-16 (2016).
- Li, D. et al. Study of antibody repertoires to the CD4 binding site of gp120 of a Chinese HIV-1-infected elite neutralizer, using 454 sequencing and single-cell sorting. Arch Virol 161, 789-799, doi:10.1007/s00705-015-2710-x (2016).
- Wu, X. et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593-1602, doi:10.1126/science.1207532 (2011).
- Diskin, R. et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334, 1289-1293, doi:10.1126/science.1213782 (2011).
- Zhou, T. et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811-817, doi:10.1126/science.1192819 (2010).
- Zhou, T. et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39, 245-258, doi:10.1016/j.immuni.2013.04.012 (2013).
- West, A. P., Jr., Diskin, R., Nussenzweig, M. C. & Bjorkman, P. J. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci U S A 109, E2083-2090, doi:10.1073/pnas.1208984109 (2012).
- Chuang, G. Y. et al. Residue-level prediction of HIV-1 antibody epitopes based on neutralization of diverse viral strains. J Virol 87, 10047-10058, doi:10.1128/JVI.00984-13 (2013).
- Liao, H. X. et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469-476, doi:10.1038/nature12053 (2013).
- Burton, D. R. et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266, 1024-1027 (1994).
- Walker, L. M. et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285-289, doi:10.1126/science.1178746 (2009).
- McLellan, J. S. et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336-343, doi:10.1038/nature10696 (2011).
- Mouquet, H. & Nussenzweig, M. C. Polyreactive antibodies in adaptive immune responses to viruses. Cell Mol Life Sci 69, 1435-1445, doi:10.1007/s00018-011-0872-6 (2012).
- Pancera, M. et al. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol 20, 804-813, doi:10.1038/nsmb.2600 (2013).
- Wardemann, H. et al. Predominant autoantibody production by early human B cell precursors. Science 301, 1374-1377, doi:10.1126/science.1086907 (2003).
- Pejchal, R. et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334, 1097-1103, doi:10.1126/science.1213256 (2011).
- Trkola, A. et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol 70, 1100-1108 (1996).
- West, A. P., Jr. et al. Design and expression of a dimeric form of human immunodeficiency virus type 1 antibody 2G12 with increased neutralization potency. J Virol 83, 98-104, doi:10.1128/JVI.01564-08 (2009).
- Mouquet, H. et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 109, E3268-3277, doi:10.1073/pnas.1217207109 (2012).
- Julien, J. P. et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog 9, e1003342, doi:10.1371/journal.ppat.1003342 (2013).
- Loos, A. et al. Glycan modulation and sulfoengineering of anti-HIV-1 monoclonal antibody PG9 in plants. Proc Natl Acad Sci U S A 112, 12675-12680, doi:10.1073/pnas.1509090112 (2015).
- Bonsignori, M. et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol 85, 9998-10009, doi:10.1128/JVI.05045-11 (2011).
- Julien, J. P. et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A 110, 4351-4356, doi:10.1073/pnas.1217537110 (2013).
- Loving, R., Sjoberg, M., Wu, S. R., Binley, J. M. & Garoff, H. Inhibition of the HIV-1 spike by single-PG9/16-antibody binding suggests a coordinated-activation model for its three protomeric units. J Virol 87, 7000-7007, doi:10.1128/JVI.00530-13 (2013).
- Huang, J. et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406-412, doi:10.1038/nature11544 (2012).
- Zhu, Z. et al. Cross-reactive HIV-1-neutralizing human monoclonal antibodies identified from a patient with 2F5-like antibodies. J Virol 85, 11401-11408, doi:10.1128/JVI.05312-11 (2011).
- Gohain, N. et al. Molecular basis for epitope recognition by non-neutralizing anti-gp41 antibody F240. Sci Rep 6, 36685, doi:10.1038/srep36685 (2016).
- Voss, J. E. et al. Reproducing SIVDeltanef vaccine correlates of protection: trimeric gp41 antibody concentrated at mucosal front lines. AIDS 30, 2427-2438, doi:10.1097/QAD.0000000000001199 (2016).
- McFarren, A. et al. A fully human antibody to gp41 selectively eliminates HIV-infected cells that transmigrated across a model human blood brain barrier. AIDS 30, 563-572, doi:10.1097/QAD.0000000000000968 (2016).
- Haynes, B. F. et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308, 1906-1908, doi:10.1126/science.1111781 (2005).
- Finton, K. A. et al. Autoreactivity and exceptional CDR plasticity (but not unusual polyspecificity) hinder elicitation of the anti-HIV antibody 4E10. PLoS Pathog 9, e1003639, doi:10.1371/journal.ppat.1003639 (2013).
- Yang, G. et al. Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. J Exp Med 210, 241-256, doi:10.1084/jem.20121977 (2013).
- Klein, J. S. et al. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc Natl Acad Sci U S A 106, 7385-7390, doi:10.1073/pnas.0811427106 (2009).
- Alam, S. M. et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci U S A 106, 20234-20239, doi:10.1073/pnas.0908713106 (2009).
- Chen, Y. et al. Common tolerance mechanisms, but distinct cross-reactivities associated with gp41 and lipids, limit production of HIV-1 broad neutralizing antibodies 2F5 and 4E10. J Immunol 191, 1260-1275, doi:10.4049/jimmunol.1300770 (2013).
- Doyle-Cooper, C. et al. Immune tolerance negatively regulates B cells in knock-in mice expressing broadly neutralizing HIV antibody 4E10. J Immunol 191, 3186-3191, doi:10.4049/jimmunol.1301285 (2013).
- Irimia, A. et al. Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design. PLoS Pathog 13, e1006212, doi:10.1371/journal.ppat.1006212 (2017).
- Verkoczy, L. et al. Induction of HIV-1 broad neutralizing antibodies in 2F5 knock-in mice: selection against membrane proximal external region-associated autoreactivity limits T-dependent responses. J Immunol 191, 2538-2550, doi:10.4049/jimmunol.1300971 (2013).
- Alsahafi, N. et al. Nef Proteins from HIV-1 Elite Controllers Are Inefficient at Preventing Antibody-Dependent Cellular Cytotoxicity. J Virol 90, 2993-3002, doi:10.1128/JVI.02973-15 (2015).
- Lynch, R. M. et al. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci Transl Med 7, 319ra206, doi:10.1126/scitranslmed.aad5752 (2015).
- Freund, N. T. et al. A New Glycan-Dependent CD4-Binding Site Neutralizing Antibody Exerts Pressure on HIV-1 In Vivo. PLoS Pathog 11, e1005238, doi:10.1371/journal.ppat.1005238 (2015).
- Friedrichs, I., Buus, C., Berger, A., Keppler, O. T. & Rabenau, H. F. Evaluation of two HIV antibody confirmatory assays: Geenius HIV1/2 Confirmatory Assay and the recomLine HIV-1 & HIV-2 IgG Line Immunoassay. J Virol Methods 224, 91-94, doi:10.1016/j.jviromet.2015.08.015 (2015).
- Gohain, N. et al. Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1. J Virol 89, 8840-8854, doi:10.1128/JVI.01232-15 (2015).
- Burton, D. R. & Mascola, J. R. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol 16, 571-576, doi:10.1038/ni.3158 (2015).
- Tiller, T. et al. Autoreactivity in human IgG+ memory B cells. Immunity 26, 205-213, doi:10.1016/j.immuni.2007.01.009 (2007).
- Klein, F. et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153, 126-138, doi:10.1016/j.cell.2013.03.018 (2013).
- Sun, Z., Lu, S., Yang, Z., Li, J. & Zhang, M. Y. Construction of a recombinant full-length membrane associated IgG library. Virus Res 238, 156-163, doi:10.1016/j.virusres.2017.06.018 (2017).
- Hoot, S. et al. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS Pathog 9, e1003106, doi:10.1371/journal.ppat.1003106 (2013).
- Victora, G. D. & Nussenzweig, M. C. Germinal centers. Annu Rev Immunol 30, 429-457, doi:10.1146/annurev-immunol-020711-075032 (2012).
- Fugmann, S. D., Lee, A. I., Shockett, P. E., Villey, I. J. & Schatz, D. G. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol 18, 495-527, doi:10.1146/annurev.immunol.18.1.495 (2000).
- Jung, D., Giallourakis, C., Mostoslavsky, R. & Alt, F. W. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol 24, 541-570, doi:10.1146/annurev.immunol.23.021704.115830 (2006).
- Martinez-Navio, J. M. et al. Host Anti-antibody Responses Following Adeno-associated Virus-mediated Delivery of Antibodies Against HIV and SIV in Rhesus Monkeys. Mol Ther 24, 76-86, doi:10.1038/mt.2015.191 (2016).
- Zinocker, S. et al. The V gene repertoires of classical and atypical memory B cells in malaria-susceptible West African children. J Immunol 194, 929-939, doi:10.4049/jimmunol.1402168 (2015).
- Lu, S., Sun, Z. & Zhang, M.-y. Generation of immortalized human naïve B cell libraries by optimized EBV transformation. Journal of Medical Discovery 2, doi:10.24262/jmd.2.1.17007 (2016).
- West, Anthony P. et al. Structural Insights on the Role of Antibodies in HIV-1 Vaccine and Therapy. Cell 156, 633-648, doi:10.1016/j.cell.2014.01.052 (2014).
- Bar, K. J. et al. Effect of HIV Antibody VRC01 on Viral Rebound after Treatment Interruption. N Engl J Med 375, 2037-2050, doi:10.1056/NEJMoa1608243 (2016).
- Kesavardhana, S. et al. Structure-based Design of Cyclically Permuted HIV-1 gp120 Trimers That Elicit Neutralizing Antibodies. J Biol Chem 292, 278-291, doi:10.1074/jbc.M116.725614 (2017).
- Cheeseman, H. M. et al. Broadly Neutralizing Antibodies Display Potential for Prevention of HIV-1 Infection of Mucosal Tissue Superior to That of Nonneutralizing Antibodies. J Virol 91, doi:10.1128/JVI.01762-16 (2017).
- Briney, B. et al. Tailored Immunogens Direct Affinity Maturation toward HIV Neutralizing Antibodies. Cell 166, 1459-1470 e1411, doi:10.1016/j.cell.2016.08.005 (2016).
- Tay, M. Z. et al. Antibody-Mediated Internalization of Infectious HIV-1 Virions Differs among Antibody Isotypes and Subclasses. PLoS Pathog 12, e1005817, doi:10.1371/journal.ppat.1005817 (2016).
- Heydarchi, B. et al. Repeated Vaccination of Cows with HIV Env gp140 during Subsequent Pregnancies Elicits and Sustains an Enduring Strong Env-Binding and Neutralising Antibody Response. PLoS One 11, e0157353, doi:10.1371/journal.pone.0157353 (2016).
- Gray, G. E., Laher, F., Lazarus, E., Ensoli, B. & Corey, L. Approaches to preventative and therapeutic HIV vaccines. Curr Opin Virol 17, 104-109, doi:10.1016/j.coviro.2016.02.010 (2016).
- Gunn, B. M. et al. Enhanced binding of antibodies generated during chronic HIV infection to mucus component MUC16. Mucosal Immunol 9, 1549-1558, doi:10.1038/mi.2016.8 (2016).
- Zhang, M.-Y. et al. Non-HIV-derived (Poly)peptides as Primary Immunogens Followed by Envelope Boost Elicit Cross-clade Neutralizing HIV-1 Antibodies. HIV Research for Prevention 2014: AIDS Vaccine, Microbicide and ARV-based Prevention Science Cape Town, South Africa, 82 (2014).
- Yuan, T., Li, J. & Zhang, M. Y. A single mutation turns a non-binding germline-like predecessor of broadly neutralizing antibody into a binding antibody to HIV-1 envelope glycoproteins. MAbs 3, 402-407 (2011).
- Bradley, T. et al. HIV-1 Envelope Mimicry of Host Enzyme Kynureninase Does Not Disrupt Tryptophan Metabolism. J Immunol 197, 4663-4673, doi:10.4049/jimmunol.1601484 (2016).
- Sok, D. et al. Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science 353, 1557-1560, doi:10.1126/science.aah3945 (2016).
- Gach, J. S. et al. HIV-1-Specific Antibody Response and Function after DNA Prime and Recombinant Adenovirus 5 Boost HIV Vaccine in HIV-Infected Subjects. PLoS One 11, e0160341, doi:10.1371/journal.pone.0160341 (2016).
- Ingale, J. et al. High-Density Array of Well-Ordered HIV-1 Spikes on Synthetic Liposomal Nanoparticles Efficiently Activate B Cells. Cell Rep 15, 1986-1999, doi:10.1016/j.celrep.2016.04.078 (2016).
- Kong, R. et al. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352, 828-833, doi:10.1126/science.aae0474 (2016).
- Mason, R. D. et al. Targeted Isolation of Antibodies Directed against Major Sites of SIV Env Vulnerability. PLoS Pathog 12, e1005537, doi:10.1371/journal.ppat.1005537 (2016).
- Behrens, A. J. et al. Composition and Antigenic Effects of Individual Glycan Sites of a Trimeric HIV-1 Envelope Glycoprotein. Cell Rep 14, 2695-2706, doi:10.1016/j.celrep.2016.02.058 (2016).
- Madani, N. et al. Antibodies Elicited by Multiple Envelope Glycoprotein Immunogens in Primates Neutralize Primary Human Immunodeficiency Viruses (HIV-1) Sensitized by CD4-Mimetic Compounds. J Virol 90, 5031-5046, doi:10.1128/JVI.03211-15 (2016).
- Caucheteux, S. M. et al. Polypropylene Sulfide Nanoparticle p24 Vaccine Promotes Dendritic Cell-Mediated Specific Immune Responses against HIV-1. J Invest Dermatol 136, 1172-1181, doi:10.1016/j.jid.2016.01.033 (2016).
- Dai, K. et al. HIV-1 Vaccine-elicited Antibodies Reverted to Their Inferred Naive Germline Reveal Associations between Binding Affinity and in vivo Activation. Sci Rep 6, 20987, doi:10.1038/srep20987 (2016).
- Huang, Y. et al. Selection of HIV vaccine candidates for concurrent testing in an efficacy trial. Curr Opin Virol 17, 57-65, doi:10.1016/j.coviro.2016.01.007 (2016).
- Tan, H. X. et al. Recombinant influenza virus expressing HIV-1 p24 capsid protein induces mucosal HIV-specific CD8 T-cell responses. Vaccine 34, 1172-1179, doi:10.1016/j.vaccine.2016.01.030 (2016).
- Patil, S. et al. Conformational Epitope-Specific Broadly Neutralizing Plasma Antibodies Obtained from an HIV-1 Clade C-Infected Elite Neutralizer Mediate Autologous Virus Escape through Mutations in the V1 Loop. J Virol 90, 3446-3457, doi:10.1128/JVI.03090-15 (2016).
- Bradley, T. et al. Structural Constraints of Vaccine-Induced Tier-2 Autologous HIV Neutralizing Antibodies Targeting the Receptor-Binding Site. Cell Rep 14, 43-54, doi:10.1016/j.celrep.2015.12.017 (2016).
- Gu, L. et al. Adenoviral vectors elicit humoral immunity against variable loop 2 of clade C HIV-1 gp120 via “Antigen Capsid-Incorporation” strategy. Virology 487, 75-84, doi:10.1016/j.virol.2015.10.010 (2016).
- Ternette, N. et al. Defining the HLA class I-associated viral antigen repertoire from HIV-1-infected human cells. Eur J Immunol 46, 60-69, doi:10.1002/eji.201545890 (2016).
- Hamlyn, E. et al. Plasma HIV viral rebound following protocol-indicated cessation of ART commenced in primary and chronic HIV infection. PLoS One 7, e43754, doi:10.1371/journal.pone.0043754 (2012).
- Igarashi, T. et al. Human immunodeficiency virus type 1 neutralizing antibodies accelerate clearance of cell-free virions from blood plasma. Nat Med 5, 211-216, doi:10.1038/5576 (1999).
- Nimmerjahn, F. & Ravetch, J. V. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol 8, 34-47, doi:10.1038/nri2206 (2008).
- Ye, Z. W. et al. Antibody-Dependent Cell-Mediated Cytotoxicity Epitopes on the Hemagglutinin Head Region of Pandemic H1N1 Influenza Virus Play Detrimental Roles in H1N1-Infected Mice. Front Immunol 8, 317, doi:10.3389/fimmu.2017.00317 (2017).
- Malbec, M. et al. Broadly neutralizing antibodies that inhibit HIV-1 cell to cell transmission. J Exp Med 210, 2813-2821, doi:10.1084/jem.20131244 (2013).
- Grant, P. M. & Zolopa, A. R. Optimal antiretroviral therapy: HIV-1 treatment strategies to avoid and overcome drug resistance. Curr Opin Investig Drugs 11, 901-910 (2010).
- Simon, V., Ho, D. D. & Abdool Karim, Q. HIV/AIDS epidemiology, pathogenesis, prevention, and treatment. Lancet 368, 489-504, doi:10.1016/S0140-6736(06)69157-5 (2006).
- Olender, S., Wilkin, T. J., Taylor, B. S. & Hammer, S. M. Advances in antiretroviral therapy. Top Antivir Med 20, 61-86 (2012).
- Vergidis, P. I., Falagas, M. E. & Hamer, D. H. Meta-analytical studies on the epidemiology, prevention, and treatment of human immunodeficiency virus infection. Infect Dis Clin North Am 23, 295-308, doi:10.1016/j.idc.2009.01.013 (2009).
- Poignard, P. et al. Neutralizing antibodies have limited effects on the control of established HIV-1 infection in vivo. Immunity 10, 431-438 (1999).
- Trkola, A. et al. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat Med 11, 615-622, doi:10.1038/nm1244 (2005).
- Mehandru, S. et al. Adjunctive passive immunotherapy in human immunodeficiency virus type 1-infected individuals treated with antiviral therapy during acute and early infection. J Virol 81, 11016-11031, doi:10.1128/JVI.01340-07 (2007).
- Klein, F. et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492, 118-122, doi:10.1038/nature11604 (2012).
- Diskin, R. et al. Restricting HIV-1 pathways for escape using rationally designed anti-HIV-1 antibodies. J Exp Med 210, 1235-1249, doi:10.1084/jem.20130221 (2013).
- Horwitz, J. A. et al. HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc Natl Acad Sci U S A 110, 16538-16543, doi:10.1073/pnas.1315295110 (2013).
- Barouch, D. H. et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature 503, 224-228, doi:10.1038/nature12744 (2013).
- Shingai, M. et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature 503, 277-280, doi:10.1038/nature12746 (2013).
- Prince, A. M. et al. Prevention of HIV infection by passive immunization with HIV immunoglobulin. AIDS Res Hum Retroviruses 7, 971-973 (1991).
- Safrit, J. T. et al. hu-PBL-SCID mice can be protected from HIV-1 infection by passive transfer of monoclonal antibody to the principal neutralizing determinant of envelope gp120. Aids 7, 15-21 (1993).
- McCoy, L. E. & Weiss, R. A. Neutralizing antibodies to HIV-1 induced by immunization. J Exp Med 210, 209-223, doi:10.1084/jem.20121827 (2013).
- Euler, Z., van Gils, M. J., Boeser-Nunnink, B. D., Schuitemaker, H. & van Manen, D. Genome-wide association study on the development of cross-reactive neutralizing antibodies in HIV-1 infected individuals. PLoS One 8, e54684, doi:10.1371/journal.pone.0054684 (2013).
- Shingai, M. et al. Most rhesus macaques infected with the CCR5-tropic SHIV(AD8) generate cross-reactive antibodies that neutralize multiple HIV-1 strains. Proc Natl Acad Sci U S A 109, 19769-19774, doi:10.1073/pnas.1217443109 (2012).
- Walker, L. M. et al. Rapid development of glycan-specific, broad, and potent anti-HIV-1 gp120 neutralizing antibodies in an R5 SIV/HIV chimeric virus infected macaque. Proc Natl Acad Sci U S A 108, 20125-20129, doi:10.1073/pnas.1117531108 (2011).
- Doria-Rose, N. A. et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509, 55-62, doi:10.1038/nature13036 (2014).
- Cortez, V., Odem-Davis, K., McClelland, R. S., Jaoko, W. & Overbaugh, J. HIV-1 superinfection in women broadens and strengthens the neutralizing antibody response. PLoS Pathog 8, e1002611, doi:10.1371/journal.ppat.1002611 (2012).
- Powell, R. L., Kinge, T. & Nyambi, P. N. Infection by discordant strains of HIV-1 markedly enhances the neutralizing antibody response against heterologous virus. J Virol 84, 9415-9426, doi:10.1128/JVI.02732-09 (2010).
- Burton, D. R. et al. A Blueprint for HIV Vaccine Discovery. Cell Host Microbe 12, 396-407, doi:10.1016/j.chom.2012.09.008 (2012).
- Zolla-Pazner, S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat Rev Immunol 4, 199-210, doi:10.1038/nri1307 (2004).
- Haynes, B. F., Kelsoe, G., Harrison, S. C. & Kepler, T. B. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol 30, 423-433, doi:10.1038/nbt.2197 (2012).
- Kwong, P. D. & Mascola, J. R. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37, 412-425, doi:10.1016/j.immuni.2012.08.012 (2012).
- Alam, S. M. et al. Differential reactivity of germ line allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation. J Virol 85, 11725-11731, doi:10.1128/JVI.05680-11 (2011).
- Jardine, J. et al. Rational HIV Immunogen Design to Target Specific Germline B Cell Receptors. Science, doi:10.1126/science.1234150 (2013).
- Alam, S. M. et al. Recognition of synthetic glycopeptides by HIV-1 broadly neutralizing antibodies and their unmutated ancestors. Proc Natl Acad Sci U S A 110, 18214-18219, doi:10.1073/pnas.1317855110 (2013).
- McGuire, A. T. et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med 210, 655-663, doi:10.1084/jem.20122824 (2013).
- Batista, F. D. & Neuberger, M. S. Affinity dependence of the B cell response to antigen: a threshold, a ceiling, and the importance of off-rate. Immunity 8, 751-759 (1998).
- Foote, J. & Milstein, C. Kinetic maturation of an immune response. Nature 352, 530-532, doi:10.1038/352530a0 (1991).
- Yang, Z. et al. Identification of Non-HIV Immunogens That Bind to Germline b12 Predecessors and Prime for Elicitation of Cross-clade Neutralizing HIV-1 Antibodies. PLoS One 10, e0126428, doi:10.1371/journal.pone.0126428 (2015).
- Klein, F. et al. Antibodies in HIV-1 vaccine development and therapy. Science 341, 1199-1204, doi:10.1126/science.1241144 (2013).
- Liao, H. X. et al. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J Exp Med 208, 2237-2249, doi:10.1084/jem.20110363 (2011).
- Xiao, X., Chen, W., Feng, Y. & Dimitrov, D. S. Maturation Pathways of Cross-Reactive HIV-1 Neutralizing Antibodies. Viruses 1, 802-817, doi:10.3390/v1030802 (2009).
- Ahmed, T. et al. Control of HIV-1 replication in vitro by vaccine-induced human CD8(+) T cells through conserved subdominant Pol epitopes. Vaccine 34, 1215-1224, doi:10.1016/j.vaccine.2015.12.021 (2016).
- Aldovini, A. Mucosal Vaccination for Prevention of HIV Infection and AIDS. Curr HIV Res 14, 247-259 (2016).
- Andersson, A. C. et al. Effect of HIV-1 envelope cytoplasmic tail on adenovirus primed virus encoded virus-like particle immunizations. Vaccine 34, 5344-5351, doi:10.1016/j.vaccine.2016.08.089 (2016).
Copyright
© This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/